March 16, 2021 -- A new intestine-on-a-chip platform enables researchers to model host-microbiome relationships to gain new insights into the mechanisms of infectious disease in the body, according to a new study published in Frontiers in Cellular and Infectious Microbiology on March 12.
Great progress has been made in gaining understanding of host-microbiome relationships using mouse models, where the microbiota and host immune cell regulatory responses can be well characterized. However, it is difficult to identify specific cellular and molecular contributors to host immune tolerance using in vivo experiments alone.
Organ-on-a-chip (organ chip) microfluidic culture technology offers a way to recapitulate organ-level structures and functions in vitro. Organ chips can be used to interrogate cellular interactions with precision at the organ level.
Most organ chips have been used to mimic human physiology, but chips created with mouse intestinal epithelial cells could provide the benefit of being able to directly compare chip responses to results of prior in vivo animal studies, according to a team led by Dr. Donald Ingber, PhD, founding director of the Harvard Wyss Institute for Biologically Inspired Engineering.
"Biomedical research strongly depends on animal models such as mice, which undoubtedly have tremendous benefits, but do not provide an opportunity to study normal and pathological processes within a particular organ, such as the intestine, close-up and in real-time," Ingber said in a statement. "This important proof-of-concept study highlights that our engineered mouse intestine chip platform offers exactly this capability and provides the possibility to study host-microbiome interactions with microbiomes from different species under highly controllable conditions in vitro."
One such host-microbiome interaction is the immune response to pathogens. Previous studies have shown that differences in gut microbiota can contribute to species-species differences in resistance to certain pathogens -- for example, Salmonella typhimurium is more infectious in humans than in mice.
The microbiome can also protect from infection by inducing host tolerance through thickening of the intestinal mucus layer or activation of the host immune system via production of cytokines. For instance, the commensal bacteria Enterococcus faecium promotes tolerance of S. typhimurium in some species.
Engineering a mouse intestine-on-chip platform
In the current study, the Wyss researchers generated mouse organoids from the intestinal tract of mice, including the duodenum, jejunum, ileum, and colon. Organoids were enzymatically disrupted and physically dissociated before being seeded on the extracellular matrix-coated porous membrane that separates two parallel microfluidic channels of an organ chip.
The second independently perfused channel mimics the blood vasculature. The porous membrane allows for the exchange of nutrients, metabolites, and secreted molecules that intestinal epithelial cells use to communicate with vascular and immune cells.
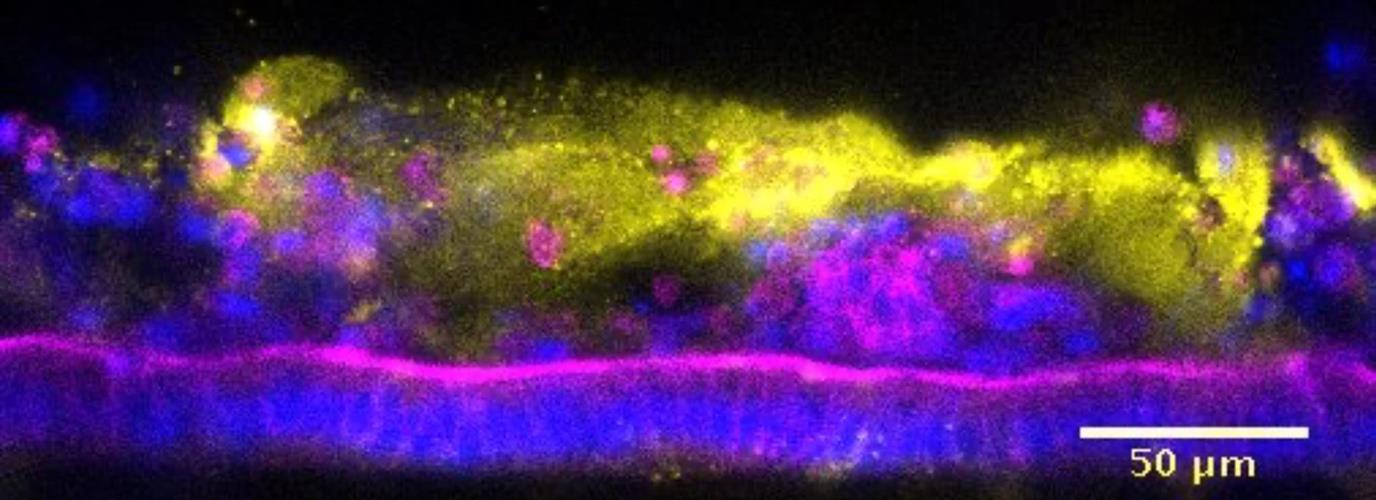
To complete the formation of region-specific intestinal chips, an organoid culture medium was then perfused through both channels. After the cells were permitted to grow for one week, the researchers confirmed that the cells formed monolayers on the chips. For this study, the team focused on the study of mouse colon chips.
"It has traditionally been extremely difficult to model host-microbiome interactions outside any organism as many bacteria are strictly anaerobic and die in normal atmospheric oxygen conditions. Organ chip technology can recreate these conditions, and it is much easier to obtain primary intestinal and immune cells from mice than having to rely on human biopsies," said first author Francesca Gazzaniga, PhD, a postdoctoral fellow at the Wyss Institute.
Gaining insights into pathogen response
The researchers sought to investigate if both individual types of bacteria and complex species-specific microbiota could be cultured on their colon chips. First, to determine if the mouse organ chips could be used to interrogate responses to a specific pathogen, the team compared the responses to co-culture with S. typhimurium to commensal bacteria from healthy mice.
The team found that S. typhimurium caused disruption of tight junctions and decreased mucus accumulation in the epithelial cells. Moreover, they found that S. typhimurium infection induced more than a five-fold increase in C-X-C motif chemokine ligand 1 (CXCL1), the mouse homolog to the human cytokine interleukin 8, production in the colon chip 24 hours after infection.
In a separate experiment, when challenged with S. typhimurium, the mouse colon chips responded differently depending on whether they were colonized with a human versus mouse complex microbiome. The researchers found variation in the microbiome composition and the extent of epithelial damage due to the pathogen between chips.
The authors noted that future studies exploring differences among microbiome compositions could help to identify bacteria that promote epithelial integrity, with potential clinical applications in microbiome-based therapies.
In this process, they also identified E. faecium isolated from the human microbiome as a bacterium that promotes host tolerance to infection. The authors noted that this bacterium also induced small regions of epithelial detachment, highlighting the potential of a commensal bacterial to have both positive and negative properties.
"Using 16s sequencing gave us a good sense of the microbial compositions of the two consortia, and high numbers of one individual species, Enterococcus faecium, generated by only one of them in the colon chip, allowed the intestinal tissue to better tolerate the infection," Gazzaniga said. "This nicely confirmed past findings and validated our approach as a new discovery platform that we can now use to investigate the mechanisms that underlie these effects as well as the contribution of vital immune cell contributions to host-tolerance, as well as infectious processes involving other pathogens."
Overall, the mouse intestine chips were found to be a useful tool that can be used in the future to study interactions between specific microbes and the intestinal epithelium. The researchers believe that this in vitro approach could also be used to identify tolerance-enhancing bacteria for use in future therapies, which may circumvent the problem of increasing antimicrobial resistance to pathogenic bacterial strains.
"The beauty of this system is that essentially all parameters you wish to study are controllable and can easily be monitored," said author Dr. Dennis Kasper, professor of medicine and immunology at Harvard Medical School. "This system is a very useful step forward."
Do you have a unique perspective on your research related to drug development or cell biology? Contact the editor today to learn more.
Copyright © 2021 scienceboard.net