September 21, 2020 -- The SARS-CoV-2 spike protein can adopt at least 10 sequential structural conformations when in contact with the human angiotensin-converting enzyme 2 (ACE2) receptor, according to a study conducted by researchers from the Francis Crick Institute and published in Nature on September 17.
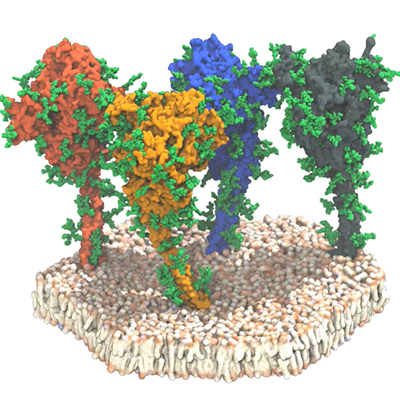
Previous studies have demonstrated that the spike trimer protein consists of a central helical stalk, made of three interacting S2 subunit components, which is covered at the top by the S1 subunit. Each S1 component contains a large N-terminal domain and a receptor binding domain.
In viral membranes the spike proteins exist in the closed form, where the RBDs are inaccessible. However, in the open conformation, one S1 subunit is opened to expose one RBD for binding with ACE2.
New insights into the specific mechanism of infection of SARS-CoV-2 will equip research groups with the understanding needed to develop efficacious vaccines and treatments.
To conduct their examination, the researchers mixed ectodomains of spike proteins with ectodomains of ACE2 prior to freezing the samples in liquid ethane for inspection by cryo-electron microscopy.
From the tens of thousands of high-resolution images obtained, the researchers identified 10 distinct spike-ACE2 complexes, ranging from tightly closed, unbound trimers to open trimers forming complexes with three ACE2 molecules. Of all the spike trimers analyzed, approximately two-thirds have an ACE2 bound. Of the unbound spike proteins, most are in the closed conformation.
Of the ACE2-bound spike trimers, around 50% accommodate one ACE2 receptor, most with a single RBD occupied at various tilts along the axis of the trimer. In these cases, the other two RBDs that are not engaged are either both closed, or one remains closed and one is in the open conformation. In the population of spike trimers, the researchers also observed 14% of trimers had two ACE2 bound and 3% had three receptors bound.
"By examining the binding event in its entirety, we've been able to characterize spike structures that are unique to SARS-CoV-2, we can see that as the spike becomes more open, the stability of the protein will reduce, which may increase the ability of the protein to carry out membrane fusion, allowing infection," said co-author Donald Benton, PhD, postdoctoral training fellow in the Structural Biology of Disease Processes Laboratory at the institute, in a statement.
Successive steps from the unbound trimer to the fully open, three-ACE2-bound trimer results in structural changes that prime the spike protein for additional binding. Once the spike is bound to ACE2 at all three of its binding sites, its central core becomes exposed to facilitate membrane fusion via the N-terminal portion of the heptad repeat 1 (HR1) region of the S2 subunit. This cooperative displacement of S1 subunits primes the rearrangements in S2 that occur in the process of membrane fusion, allowing infection to proceed.
The researchers hope that the more that's understood about how SARS-CoV-2 differs from other coronaviruses, the more targeted the development of new treatments and vaccines will become. The team is continuing their work in examining spike structures to better understand the mechanisms of viral infection and evolution.
"There's so much we still don't know about SARS-CoV-2, but its basic biology contains the clues to managing this pandemic," said Steve Gamblin, PhD, group leader at the Francis Crick Institute's Structural Biology of Disease Processes Laboratory. "By understanding what makes this virus distinctive, researchers could expose weaknesses to exploit."
Do you have a unique perspective on your research related to infectious disease research or virology? Contact the editor today to learn more.
Copyright © 2020 scienceboard.net