February 19, 2020 -- Critical research reveals the 3D atomic-scale map of the spike protein of the 2019 novel coronavirus (2019-nCoV), which is responsible for infecting humans. This breakthrough will aid in the development of vaccines and antiviral drugs to combat the spread of the virus. The research was published in Science on February 19.
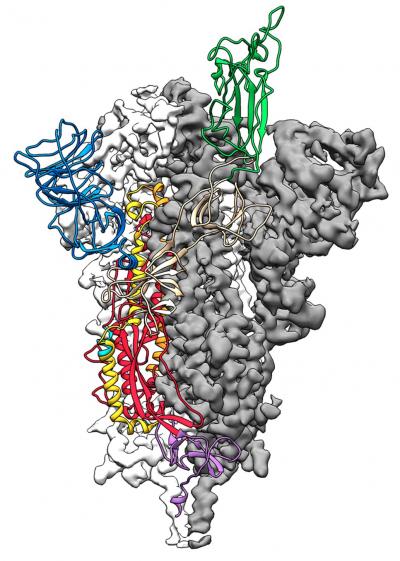
2019-nCoV makes use of a densely glycosylated spike (S) protein to gain entry into host cells. The S protein is a trimeric class I fusion protein that undergoes conformational rearrangement to fuse the viral membrane with the host-cell membrane. Due to the indispensable function of the S protein in viral infection, it represents a target for antibody-mediated neutralization. Therefore, characterization at the atomic level would guide vaccine design and development.
Applying stabilization methods that have been previously validated on other coronaviruses, researchers from the University of Texas (UT) at Austin stabilized the structure of the 2019-nCoV S protein, allowing them to visualize the domain organization of the protein. Next, they used cryogenic electron microscopy (cryo-EM) to reveal particle density and obtain 3.5 Å resolution 3D reconstruction of the asymmetrical trimer. The researchers observed the protein in both conformational arrangements and noted that the phenomenon may be conserved among Coronaviridae, in which receptor-binding leads to a conformational change.
"As soon as we knew this was a coronavirus, we felt we had to jump at it because we could be one of the first ones to get this structure," said Jason McLellan, PhD, associate professor at UT Austin, in a statement. "We knew exactly what mutations to put into this because we've already shown these mutations work for a bunch of other coronaviruses."
The research team has dedicated many years to studying other coronaviruses, including severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV). Specifically, they developed methods for analyzing the molecular structure of spike proteins to make it easier to search for vaccine candidates. This gave them an advantage when it came to studying 2019-nCoV. Based on their analysis, the overall structure of the 2019-nCoV S protein resembles that of the SARS-CoV S protein with only minor differences.
Indeed, just two weeks after receiving the genomic sequence of the virus from Chinese researchers, McLellan and colleagues designed and produced a sample of their stabilized spike protein. Then, after another 12 days, they were able to reconstruct the 3D atomic map of the protein and draft a manuscript for submission and expedited peer review.
The team found that 2019-nCoV has a high affinity for binding to angiotensin-converting enzyme 2 (ACE2), which may explain why it can easily spread from human to human. Knowing the atomic-level structure of the 2019-CoV spike protein will allow for additional protein engineering efforts to improve antigenicity and protein expression for vaccine development. Furthermore, the atomic-level detail will enable the design and screening of small molecules with fusion-inhibiting potential.
Next, the researchers plan to use their molecule to pursue another line of attack against the virus by using the molecule as a "probe" to isolate naturally produced antibodies from patients who have been infected with the 2019-nCoV and successfully recovered. In large enough quantities, these antibodies could help treat a coronavirus infection soon after exposure. For example, the antibodies could protect soldiers or healthcare workers sent into an area with high infection rates on too short notice for the immunity from a vaccine to take effect.
Do you have a unique perspective on your research related to virology and infectious disease research? Contact the editor today to learn more.
Copyright © 2020 scienceboard.net






