February 8, 2021 -- Programmable medicines that can be controlled by synthetic genetic components are not yet a clinical reality. But synthetic components can now be reconfigured so they don't overwhelm host cells, moving the technology a step closer to clinical reality, according to new research published February 8 in Nature Communications.
Synthetic biology is an interdisciplinary research field that uses engineering principles to create biological components that don't exist in the natural world. These synthetic components mimic naturally evolved organisms but are customized to diagnose and treat diseases, including cancer.
For example, synthetic gene circuits are an application of synthetic biology that control gene expression through the use of DNA-encoded tools that are delivered to predefined sites in the body. Sophisticated gene circuits, such as positive feedback loops, can be broken down into small modules with reduced complexity.
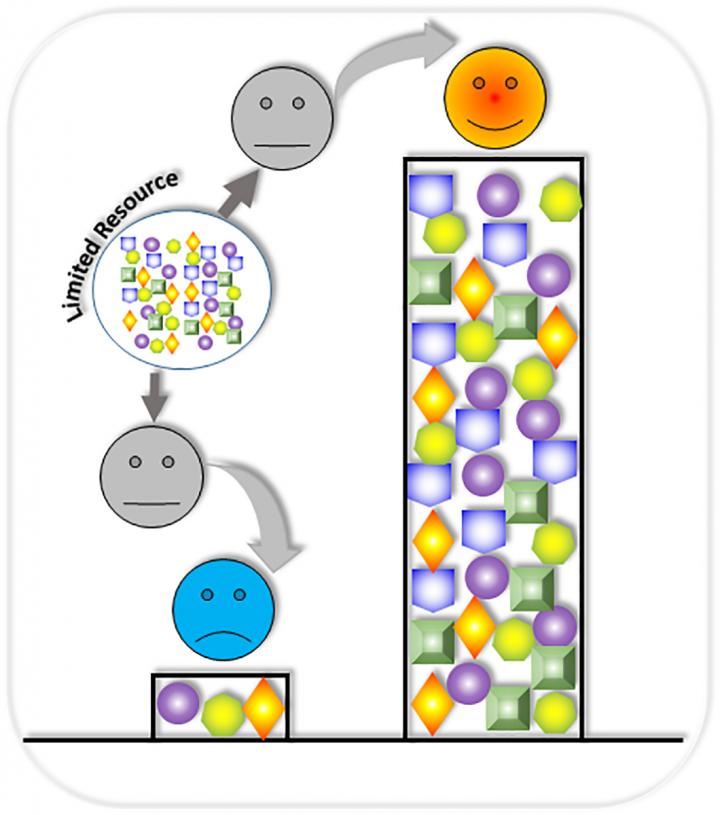
However, when modules are assembled, the whole circuit often does not function as expected. This high rate of failure can be attributed, in part, to competition over shared gene expression resources, which are limited. These include cellular resources in the host cell, such as transcriptional and translational machinery (RNA polymerases and ribosomes). Resource competition within gene circuits may change overall circuit behaviors.
"The circuits in the chain are designed to perform different functions, but they must compete with each other for the cell's limited resources," explained lead author Xiaojun Tian, PhD, assistant professor at the Arizona State University School of Biological and Health Systems Engineering, in a statement. "We would find circumstances where one gene circuit in a chain would consume 90 percent of a host cell's available resources, leaving only 10 percent for the remaining circuit."
To address this limitation, the team built a synthetic cascading bistable switches (Syn-CBS) circuit in a single strain with two coupled self-activation modules to achieve two successive cell fate transitions (desired control of gene expression). The first module was designed with self-activation of AraC, part of the L-arabinose operon that breaks down five-carbon sugars, which is controlled by Arabinose (L-ara) in the genome of Escherichia coli. The other module was designed with self-activation of LuxR, a DNA-binding protein, and is controlled by the quorum-sensing signal 3oxo-C6-HSL (C6). The switches were tagged with fluorescent proteins to indicate activation.
The researchers found that activation of one of the switches acts against the activation of the other switch, showing a negative inverse relationship. This is because the two modules compete for limited resources and thus inhibit each other indirectly. Moreover, the relative strength of connections between the modules determines the winner of resource competition in the circuit.
To further study the relationship, they measured cell fate transitions at the single-cell level with flow cytometry under various concentrations of L-ara. Here, the team found that when activation was induced for the first module, cells were not activated for the second module.
To decouple the resource competition, the researchers studied the behaviors of the two separated bistable switches (Syn-SBS) system. By applying various levels of inducers in the system, the researchers found that when the first module is activated quickly, it represses the activation of the second module. Under moderate inducement, the activation speeds of both modules are similar, leading to coactivation.
However, when the second module is quickly activated, it completely blocks the activation of the first module. This results in winner-take-all behavior in which the first activated switch always suppresses the activation of the second switch.
To attempt to achieve costimulation and remove undesired resource competition, the team constructed a two-strain Syn-CBS circuit by dividing the two modules into two separate cells, instead of placing one whole gene circuit in a single cell. They observed stable coexistence of the two populations of activated cell states within the separate cells. The authors suggested that the adverse effects of resource competition are minimized through a division of labor among different microbial species.
"Instead of dividing resources, each cell can perform 100 percent of its assigned workload," said Tian. "The host cells perform as a connected unit without depleting any one cell's resources - and each gene circuit becomes a winner."
Strategies to improve resource allocation of host cells can influence and improve the performance of gene circuits. The authors noted that the two-strain Syn-CBS circuits can be used to study new ways to dynamically deliver multiple programmable medicines.
"There are many different kinds of cells in a cancer mass," said Tian. "Some cells are responsive to chemotherapy and others are not, causing treatment resistance. New multitasking synthetic gene circuitry configuration can be constructed to prevent the cells from metastasizing in the first place, while simultaneously making them more receptive to treatment."
Do you have a unique perspective on your research related to synthetic biology? Contact the editor today to learn more.
Copyright © 2021 scienceboard.net