February 4, 2021 -- Researchers have identified a pattern of deletions in the spike glycoprotein of the SARS-CoV-2 virus that can prevent antibody binding, which is part of the body's immune response to a foreign virus. These deletions occur as a recurring pattern of evolution, according to a study published in Science on February 3.
Coronaviruses, such as SARS-CoV-2, acquire substitutions due to a proofreading RNA-dependent RNA polymerase. SARS-CoV-2 can also evade immune responses by selectively deleting small portions of its genetic material.
However, RNA-dependent RNA polymerase proofreading cannot correct deletions. This adaptive evolution of the spike protein is perpetuated by a tolerance for deletions.
Some of these deletions occupy defined antibody epitopes within the amino (N)-terminal domain (NTD) and deletions at multiple sites can confer resistance to neutralizing antibodies.
"You can't fix what's not there," said senior author Paul Duprex, PhD, director of the Center for Vaccine Research at the University of Pittsburgh, in a statement. "Once it's gone, it's gone, and if it's gone in an important part of the virus that the antibody 'sees,' then it's gone for good."
Finding a pattern of deletions
Often, emerging viruses result in pandemics due to the introduction of antigenic novelty in a virus that enables it to avoid host immune responses, as well as enabling the reinfection of previously immune individuals. The authors of the current study suggested that deletions represent a potential mechanism through which the SARS-CoV-2 spike protein rapidly acquires genetic and antigenic novelty.
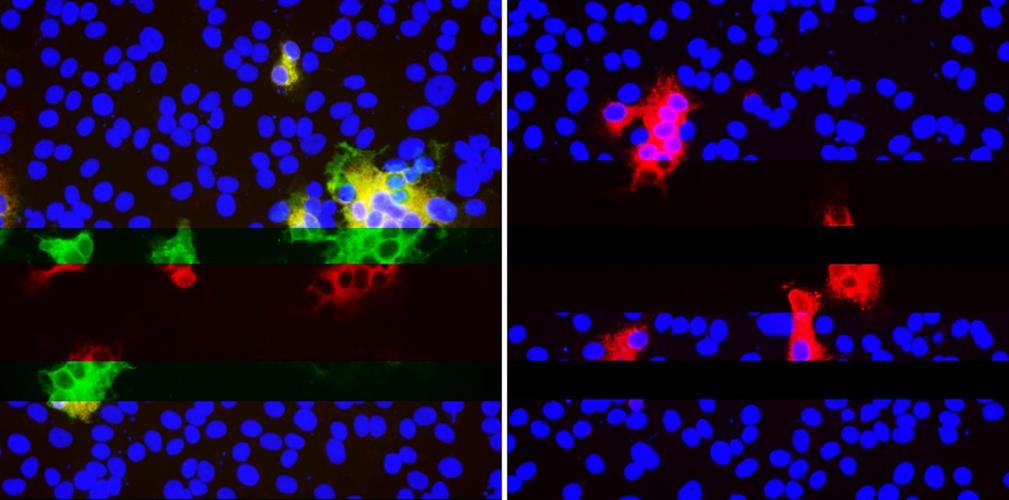
The University of Pittsburgh group first came across these neutralization-resistant deletions in a sample from an immunocompromised patient, who was infected with SARS-CoV-2 for 74 days before ultimately dying from COVID-19. This gave the virus ample time to mutate, according to the authors. They hypothesized that the evolutionary pattern defined by recurrent deletions alters defined antibody epitopes.
The researchers interrogated patient metadata sequences from the Global Initiative on Sharing All Influenza Data (GISAID). They were searching for similar viruses and identified eight patients with deletions in the spike protein of samples longitudinally over a period of weeks to months. During the early time point, each patient had intact spike sequences and then at later time points had deletions within the spike gene. The team suggests that the deletions arose independently due to a common selective pressure to produce convergent outcomes.
Searching for more deletions, the researchers identified 1,108 viruses with deletions in the spike gene from a dataset of 146,795 sequences (deposited from December 2019 to October 2020). They found that 90% of the deletions occurred in four sites -- called recurrent deletion regions (RDRs)-- within in the NTD. In all, 93% of the deletions were in frame (a deletion of at least three bases that removes an entire codon), did not produce a stop codon, and maintained the open reading frame of the virus.
"Evolution was repeating itself," said lead author Kevin McCarthy, PhD, assistant professor of molecular biology and molecular genetics at the University of Pittsburgh. "By looking at this pattern, we could forecast. If it happened a few times, it was likely to happen again."
Learning more about SARS-CoV-2 recurrent deletion regions
The RDRs identified in the group's analysis formed four distinct lineages/branches. The deletion lengths and positions vary, including loss of spike glycoprotein residues 144/145 in RDR2, loss of residues 243-244 in RDR4, and loss of residues 69-70 in RDR1. Alternatively, the RDR3 largely consists of three nucleotide deletions in codon 220. Moreover, variants of RDR1 and RDR3 are polarized to specific clades and geographies. This is likely the result of successful lineages circulating in regions with strong sequencing initiatives.
Recurrence and convergence of RDR deletions, especially during long-term infections, is indicative of adaptations in response to a common selective pressure, noted the authors. In these cases, the deletions occupy two distinct surfaces on the spike glycoprotein NTD. Both sites contain antibody epitopes. For instance, the epitope for a neutralizing antibody, 4A8 that binds the NTD, is formed by the beta sheets and extended connecting loops that hold RDR2 and RDR4.
The researchers generated a panel of spike protein mutants expressing the four RDRs to assess the effect that deletions have on expression and antibody binding. Vero E6 cells were transfected with plasmids expressing the mutant glycoproteins, and indirect immunofluorescence was used to determine if RDR deletions modulated 4A8 binding.
As expected, RDR1 and RDR3 had no effect on the binding of the monoclonal antibody, confirming they alter different sites. Alternatively, RDR2 and RDR4 deletions completely abolished binding of 4A8 while still allowing a monoclonal antibody to bind to the receptor binding domain. The researchers demonstrated that in vivo, the deletion variants can resist neutralization by monoclonal antibodies.
The data also showed that naturally arising and circulating variants of SARS-CoV-2 have altered antigenicity. In vitro, in the presence of immune serum, deletion variants acquire resistance to NTD directed antibodies by not RBD antibodies. Furthermore, they found no major difference in neutralization between spike-deleted and undeleted viruses, suggesting that many more changes would be required to generate serologically distinct SARS-CoV-2 variants.
"Going after the virus in multiple different ways is how we beat the shapeshifter," said Duprex. "Combinations of different antibodies, combinations of nanobodies with antibodies, different types of vaccines. If there's a crisis, we'll want to have those backups."
The authors noted that their analysis preceded the description of currently relevant lineages of SARS-CoV-2, including the U.K. strain (B.1.1.7). Their survey of deletion variants identified the first representative of what would become the B.1.1.7 lineage.
"These real-world outcomes demonstrate the predictive potential of this and like approaches and show the need to monitor viral evolution carefully and continually," according to the authors.
Do you have a unique perspective on your research related to virology or immunology? Contact the editor today to learn more.
Copyright © 2021 scienceboard.net