August 4, 2021 -- A new tool called protein-protein interaction sequencing (PROPER-seq) allows researchers to map complex networks of proteins within cells of interest without the need for specialized resources, like antibodies or premade gene libraries. The methodology was published in Molecular Cell on August 3.
Proteins are long strands of amino acids that are essential cellular components, providing structure, function, and regulation of the body's tissues and organs. Proteins serve not only individual purposes but also interact with other proteins to carry out complex functions through protein-protein interactions (PPIs). All of the PPIs in a cell form a PPI network.
Traditionally, identification of PPI has been a time-consuming and expensive endeavor. Now, bioengineers from the University of California, San Diego, led by professor Sheng Zhong, PhD, have developed a technology capable of revealing PPIs among thousands of proteins in a single experiment.
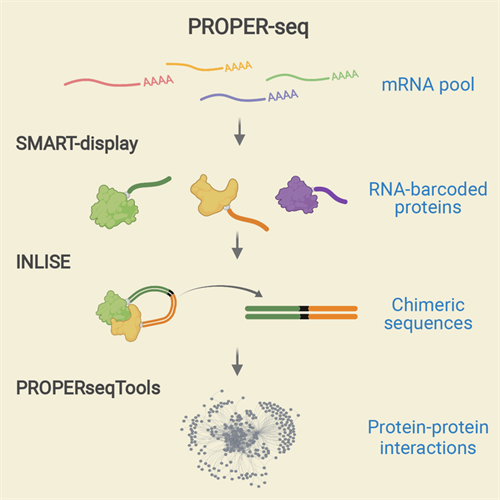
The PROPER-seq assay labels every PPI with a unique RNA sequence, which can be read by DNA sequence labels through next-generation sequencing. To do this, Zhong's team developed a new technique called SMART-display, which attaches a unique RNA barcode to every protein. This process is followed by a method called incubation, ligation, and sequencing (INLISE) that converts two attached, barcoded, interacting proteins into chimeric DNA sequences. The last component of the system is a software package called PROPERseqTools, which incorporates statistical tools to identify the PPIs from the DNA sequencing data.
Validating the PROPER-seq assay
The researchers applied PROPER-seq on human embryonic kidney cells (HEK293T cells), T lymphocytes (Jurkat cells), and endothelial cells (human umbilical vein cells), identifying 210,518 PPIs involving 8,635 proteins. Cumulatively, PROPER-seq expands the reference map of the human protein interactome with more than 200,000 previously uncharacterized PPIs, according to the authors.
The team created a public database with a web interface to download, search, and visualize these PPIs.
"PROPER-seq is capable of scanning the order of 10,000 x 10,000 protein pairs in one experiment," said first author Kara Johnson, PhD, a recent University of California, San Diego bioengineering alumna, in a statement.
To validate the workflow, the team compared PPIs identified by PROPER-seq to previously characterized PPIs from PPI databases. They found over 1,300 PPIs via PROPER-seq than coimmunoprecipitation experiments and more than 2,400 PPIs than affinity purification-mass spectrometry experiments.
Next, the researchers experimentally validated four newly identified PPIs that have not been previously reported in the literature. The PPIs involved poly (ADP-ribose) polymerase 1 (PARP1), an essential DNA repair protein and the target of several human cancers. The associated PPI proteins were involved in the trafficking of molecules and transcription regulation. These in vivo experiments suggest mechanistic links between PARP1 and the import/export of molecules to/from the nucleus, as well as gene transcription.
In addition, the team found that 100 PPIs overlap human synthetic lethal gene pairs. Synthetic lethal gene pairs can cause cell death when both genes of the gene pair are lost. This finding suggests a connection between physical interactions and human genetic interactions, the authors noted.
In the future, the team hopes PROPER-seq can assist researchers in screening many protein pairs and identify PPIs of interest. In addition, the PROPER-seq-identified PPIs from different labs can expand the PPI networks' reference maps and illuminate cell-type-specific PPIs.
Do you have a unique perspective on your research related to proteomics or genomics? Contact the editor today to learn more.
Copyright © 2021 scienceboard.net