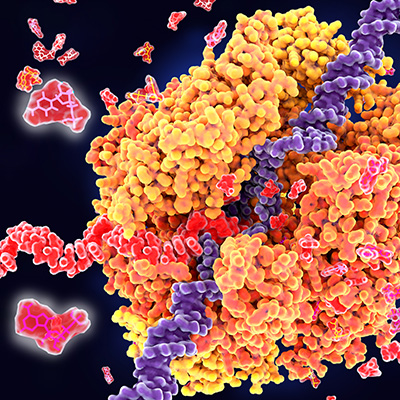
July 7, 2022 -- Researchers from the Salk Institute and Rutgers University have determined the molecular structure of HIV Pol, a protein that plays a key role in the late stages of HIV replication. Determining the molecule’s structure helped answer longstanding questions about how the protein breaks apart to advance the virus replication process.
HIV Pol breaks into three enzymes: a protease, reverse transcriptase, and integrase. Led by Dmitry Lyumkis and Eddy Arnold, the research team discovered the protease plays a critical role in initiating the replication process by chopping up the molecule to separate the other components (Science Advances, July 6, 2022, Vol. 8:27).
The protease breaks free first from the larger polyprotein HIV Gag-Pol and then from HIV Pol, initiated by self-cleaving from the rest of the molecule, aided by reverse transcriptase and, possibly, integrase, the study team said.
The researchers used cryogenic electron microscopy to reveal the 3D structure of the HIV pol protein molecule, which led to the discovery that Pol is a dimer. The finding was a surprise because other similar viral proteins are single-protein assemblies, according to the researchers.
In the two-sided structure, the protease component of Pol is "loosely tethered" to the reverse transcriptase component in a binding configuration that keeps the protease slightly flexible, meaning the protease can move and in turn initiates the cutting of polyproteins that is a prerequisite for viral maturation.
Current HIV treatments include multiple classes of inhibitors for all three enzymes, but this discovery reveals a new vulnerability that could be targeted with drugs. More research is needed that studies the structure of the larger and more complex polyprotein Gag-Pol and the role of integrase in assembling the mature form of the HIV virus during replication, the researchers added.
Copyright © 2022 scienceboard.net