December 19, 2019 -- Researchers from Columbia University capture de novo images that reveal unexpected presence transposition proteins that are essential for interactions of CRISPR-like cascade complex. The study was published in Nature on December 18.
CRISPR with a twist
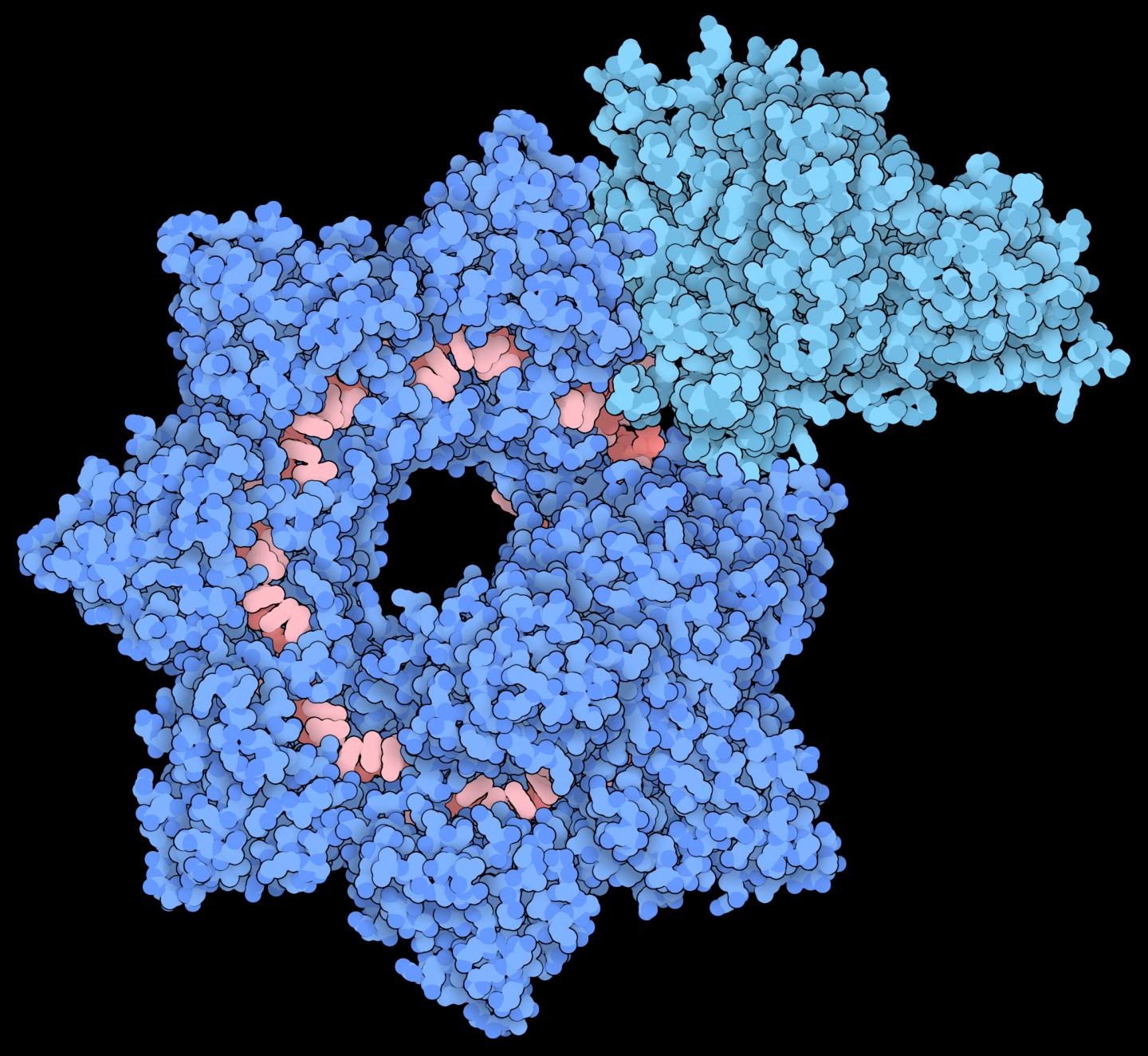
The research team used cryo-electron microscopy to generate de novo resolution models to study the "insertion of transposable elements by guide RNA-assisted targeting" (INTEGRATE) system. Transposon Tn6677 was derived from V. cholerae, in a system that was previously thought to be homologous to CRISPR systems in E. coli. The new structural models showed that this complex has two sections arranged in a helical filament. The larger section, called Cascade, holds guide RNA which is used to scan for matching DNA sequences. Once a target is located and bound, the smaller section, called TniQ, recruits additional enzymes to modify the DNA. This transposition protein dimerizes with the Cascade when DNA is bound.
The scanning function of the INTEGRATE system is similar to traditional CRISPR systems, in that a Cascade complex carries guide RNA. However, INTEGRATE is able to target DNA for highly accurate insertion of genetic payloads. The researchers had previously proposed that the INTEGRATE system uses transposition machinery for gene "jumping" where sequences of DNA that move (or jump) from one location in the genome to another. The current study proved their hypothesis correct.
"We showed in our first study how to leverage INTEGRATE for targeted DNA insertions in bacterial cells," says Sam Sternberg, PhD, assistant professor of biochemistry & molecular biophysics at Columbia University Vagelos College of Physicians and Surgeons. "These new images, explain the biology with incredible molecular detail and will help us improve the system by guiding protein engineering efforts."
Integrating INTEGRATE
CRISPR-Cas9 technology has swept the scientific community and is rapidly becoming a commonplace tool for researchers to make precise modifications to genomes. This complex system requires target DNA to be broken and repaired by the host cell's own machinery, making the repair process challenging to control. Moreover, existing CRISPR tools often cannot precisely insert large genetic payloads, which is often required to ensure safe gene therapies.
The new INTEGRATE system can address this challenge by accurately inserting large DNA sequencing without relying on the cell's machinery, but rather on transposition proteins. This indicates that the system could more accurately and more efficiently make gene modifications than other CRISPR systems. For example, INTEGRATE could be used in neurons where other systems have not been successful.
In the future, the research team hopes to explore how INTEGRATE provides a checkpoint while developing the tool for new therapeutic approaches to disease. This proofreading checkpoint is essential for accuracy of gene insertion. Current CRISPR systems often suffer from "off-target effects" in which unintended sequences are modified. The researchers believe that the collaboration between Cascade and TniQ in INTEGRATE may provide a viable solution for this problem.
Do you have a unique perspective on your research related to genetic engineering? Contact the editor today to learn more.
Copyright © 2019 scienceboard.net






