October 8, 2021 -- An atlas of the mouse cerebrum and primary motor cortex down to the level of single cells was published in a pair of papers on October 6 in Nature. The research reveals the mammalian brain's organization at an unprecedented level of detail and sheds new light on gene regulatory factors, promising to inform future work on developing targeted therapies for human diseases.
The mouse brain, which is the size of a pea and contains approximately 10 million neurons and glial cells, has proven to be a powerful model for studying many human brain functions, diseases, and mental disorders in part because more than 90% of human genes are shared with mice.
The two Nature papers map the organization of cells within two key regions of the mouse brain (the cerebrum and primary motor cortex, respectively) with unprecedented depth and specificity, down to its molecular and cellular components. At the same time, the papers probe the transcriptomic, epigenomic, and regulatory factors and elements that endow specific types of neurons with function and purpose.
"We want a comprehensive parts list for the brain, including not just the locations and connections of the neurons, but also the molecular and epigenetic fingerprints that give them their specialized identity," Eran Mukamel, PhD, director of the Computational Neural DNA Dynamics Lab and associate professor in the department of cognitive science at University of California, San Diego (UCSD), said in a statement.
The researchers hope this new level of detail, which was enabled by advances in imaging, sequencing, and computational technologies, will inform future work on developing targeted therapies for human diseases and disorders.
"To truly understand how the brain functions, and from that knowledge develop new drugs and therapies to improve human lives and health, we need to see and quantify brain structure, organization and function down to the level of single cells," Bing Ren, PhD, director of the Center for Epigenomics and professor of cellular and molecular medicine at UCSD School of Medicine, said. Ren is also a co-founder of Arima Genomics and Epigenome Technologies.
Mukamel and Ren are co-authors on both papers, along with colleagues from UCSD and other institutions, including the Allen Institute of Brain Sciences, the Salk Institute for Biological Studies, and John Hopkins University.
The research is a part of multiyear, multi-institutional program by the U.S. National Institutes of Health called the Brain Initiative Cell Census Network (BICCN), which aims to provide researchers and the public with a comprehensive reference of the diverse cell types in human, mouse, and nonhuman primate brains.
Mouse cerebrum regulatory factors
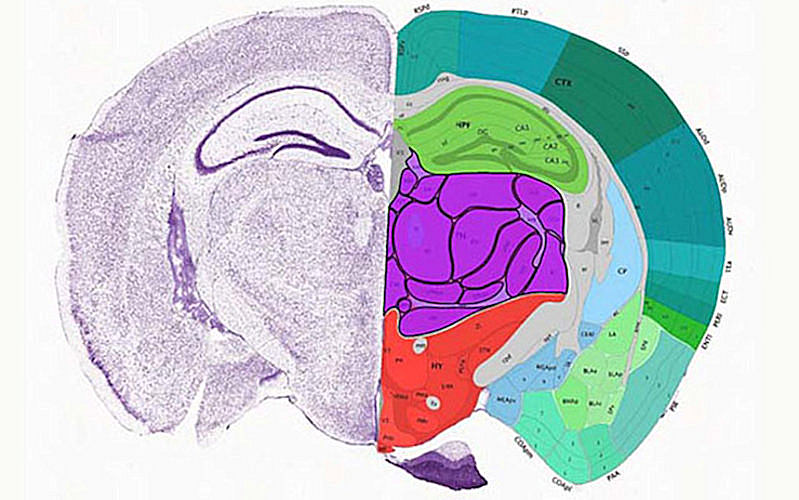
The first paper in Nature (October 6, 2021, Vol. 598, pp. 129-136) describes the creation of an atlas of gene regulatory elements in the mouse cerebrum, the evolutionarily newest region of the brain that supports high-level sensory perception, motor control, and cognitive functions.
Ren et al performed single-nucleus transposase-accessible chromatin-using sequencing (ATAC-seq) assays of more than 800,000 cells from 45 dissected regions in the adult mouse cerebrum. Next, these chromatin data were used to produce comprehensive maps of the cis-regulatory elements (CREs; regions of noncoding DNA that regulate the transcription of neighboring genes) in 160 distinct cell types.
The results showed that each of the 45 regions of the cerebrum had different types of neurons and that the specificity of their spatial distribution and function could be predicted by the unique set of CREs within each cell type. In fact, some of the cell-type-specific CREs were found to drive reporter gene expression in specific subclasses of neurons.
Intriguingly, for 69.2% of the mouse brain CREs the researchers singled out, they identified human genome sequences with high similarity, suggesting that these homologues may act as regulatory elements in humans as well.
Sequencing tech to map the primary motor cortex
The second paper (pp. 103-110), published in the same issue of Nature, describes a transcriptomic and epigenomic cell atlas of the mouse primary motor cortex, a brain region fundamental to movement.
The authors used more than 500,000 single-cell transcriptomic and epigenomic measurements compiled by BICCN to assess the molecular signatures of cell types in the primary motor cortex. The BICCN datasets comprise transcriptomic (scRNA-seq and snRNA-seq), DNA methylation (snmC-seq2), and chromatin accessibility (snATAC-seq) data.
The researchers used cutting-edge computational integration methods (specifically, the linked inference of genomic experimental relationships [LIGER] and SingleCellFusion packages) to integrate these multimodal data into a multimodal atlas of 56 neuronal cell types along with their molecular, genomic, and anatomic features.
The authors quantitatively validated the reproducibility of the 56 neuronal cell types by replicating them across multiple analysis methods, sequencing technologies, and modalities.
"Together with complementary BICCN datasets from spatial transcriptomics, connectivity, and physiology, as well as cross-species comparative studies, our results help to establish a multifaceted understanding of brain cell diversity," the authors wrote.
The datasets are available via the NeuroScience Multi-omic Data Archive (NEMO).
Do you have a unique perspective on your research related to single-cell sequencing? Contact the editor today to learn more.
Copyright © 2021 scienceboard.net