September 25, 2019 -- A new synthetic vaccine developed by researchers at the University of Bristol and the French National Centre for Scientific Research can be stored at warmer temperatures due to an engineered scaffold design. The details of the Chikungunya vaccine candidate are published in Science Advances on September 25.
Despite the advanced technologies available in the modern world, infectious diseases still affect millions worldwide. Vaccines have been instrumental in eliminated many infectious diseases, such as smallpox. However, epidemics caused by infectious disease still prevent severe challenges to certain populations, particularly in countries that do not have adequate infrastructures and resources to prevent or manage outbreaks. Recent infectious disease examples are Chikungunya and Zika which are normally transmitted by mosquitoes in sub-Saharan Africa but has spread to other parts of the world.
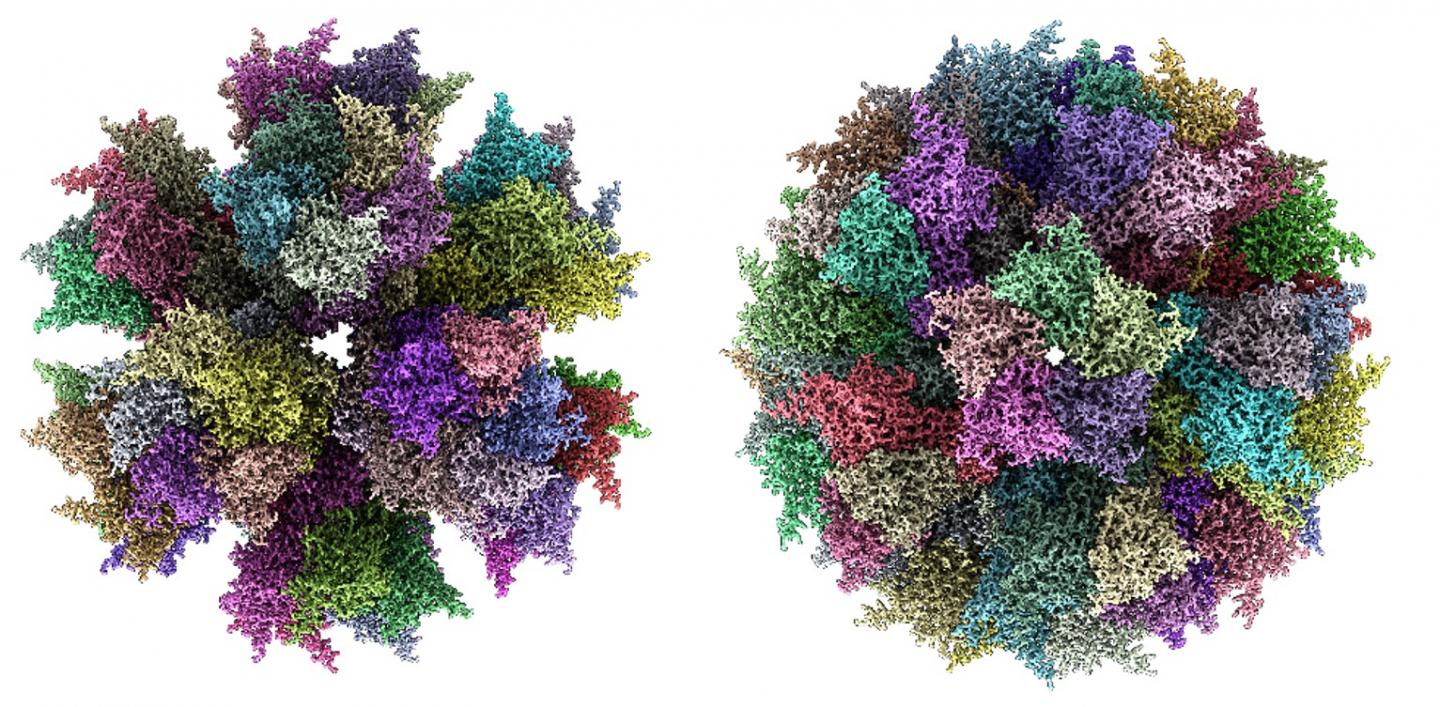
The research team created self-assembling virus-like particles called adenovirus-derived multimeric protein-based self-assembling nanoparticle scaffold (ADDomers) specifically designed to facilitate plug-and-play display from multiple epitomes of pathogens. The team started with a penton base protein and inserted DNA coded for selected epitomes into a designed insertion locus of the virus-like protein scaffold. They then tested the scaffold with a variety of epitomes from various pathogens to test capacity of insertion loci in the process.
"We were working with a protein that forms a multimeric particle resembling a virus but is completely safe, because it has no genetic material inside" said Pascal Fender, expert virologist at Centre for Scientific Research. "Completely by chance, we discovered that this particle was incredibly stable even after months, without refrigeration."
"This particle has a very flexible, exposed surface that can be easily engineered", added Imre Berger, Director of the Max Planck-Bristol Centre for Minimal Biology in Bristol. "We figured that we could insert small, harmless bits of Chikungunya to generate a virus-like mimic we could potentially use as a vaccine."
The scientists used cryo-electron microscopy to validate their technique. Cryo-EM enables researchers to determine the structure of a sample at almost atomic resolution. However, this form of microscopy generates a massive data set and requires large parallel computing power. So, the researchers partnered with computer technology giant Oracle.
With the assistance of Oracle's high-performance cloud infrastructure, they developed a novel computational approach to drafting digital models of their synthetic vaccine. The software was seamlessly integrated on the cloud to accomplish this analysis. "We were able to process the large data sets obtained by the microscope on the cloud in a fraction of the time and at much lower cost than previously thought possible" said Christopher Woods, IT specialist at the University of Bristol.
Using the data cloud for scientific discovery will allow researchers to achieve breakthroughs faster than ever before. The particles the scientists designed with on the cloud were tested in immunization studies in mice, provided promising results and the team plans to move forward in testing. This project is just one example of the technology can be integrated into the scientific discovery process. "We can now rapidly engineer similar vaccines to combat many other infectious diseases just as well" said Berger.
Do you have a unique perspective on your research related to structural biology? The Science Advisory Board wants to highlight your research. Contact the editor today to learn more.
Copyright © 2019 scienceboard.net






