May 11, 2021 -- Artificial intelligence (AI) can be used to speed up the computationally intensive process of light-field microscopy (LFM), a 3D microscopic imaging method. Researchers who developed the technique refer to it as hybrid LFM (HyLFM) and described the method in a paper published on May 7 in Nature Methods.
The approach gives biologists a promising new imaging tool for creating high-speed visualizations of dynamic processes over large, 3D fields-of-view (FOVs), such as capturing neuronal activity distributed over the entire brain.
As the name "light field" suggests, the theory underlying LFM is based on the interpretation of light as a field, much like a magnetic field, with different amounts of light flowing in every direction through every point in space. LFM works by obtaining 3D spatial information in a single camera frame, and it can take days of computation time for the vast quantities of captured data to be converted into 3D volumes and movies.
Another laboratory imaging technique, light sheet microscopy (LSM), is faster than LFM because it homes in on a single 2D plane of a given sample at one time. Because LSM only illuminates the observed plane, it is also less intrusive than LFM and is therefore favored for use in cell biology and for microscopy of in vivo organs, embryos, and organisms.
The new HyLFM approach detailed in the Nature Methods paper combines the advantages of each technique: It uses LFM to image large 3D samples and uses LSM to train an AI algorithm based on deep learning, which then creates an accurate 3D picture of the sample. The researchers, who are affiliated with institutions in Germany, Sweden, Switzerland, and Italy, summarize HyLFM as "a framework for deep learning-based microscopy with continuous ground truth generation."
"Ultimately, we were able to take 'the best of both worlds' in this approach," said co-author Nils Wagner, currently a doctoral student at the Technical University of Munich, in a statement. "AI enabled us to combine different microscopy techniques, so that we could image as fast as light-field microscopy allows and get close to the image resolution of light-sheet microscopy."
The HyLFM system uses a convolutional neural network (CNN), called HyLFM-Net, for light-field data processing and 3D image reconstruction. A novel aspect of the HyLFM system is that this CNN-driven 3D image reconstruction is enhanced on the fly by simultaneous acquisition of high-resolution image data for continuous training and validation.
This is achieved by adding a simultaneous selective-plane illumination microscopy (SPIM) modality into the LFM setup, which continuously scans through the volume at high speed with an electrically tunable lens, producing high-resolution ground truth images of single planes for validation, training, or refinement of the CNN.

This approach allows the network to be retrained when its reconstructions do not agree with the individual SPIM plane images taken during continuous validation.
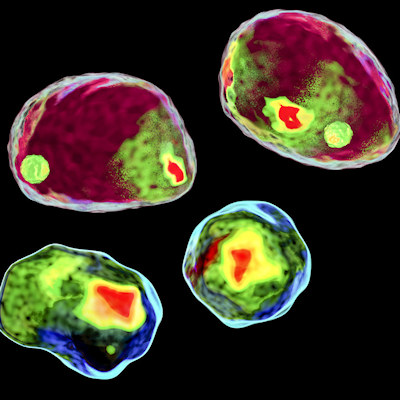
To test the ability of the HyLFM system to capture dynamic cellular movements, the researchers trained HyLFM-Net on images of the beating heart of medaka (Oryzias latipes) hatchlings (eight days postfertilization [dpf]) across a 350 × 300 × 150-µm3 FOV at a volumetric speed of 40-100 Hz. In another experiment, HyLFM-Net was trained on five dpf transgenic larval zebrafish (Danio rerio) brains expressing the nuclear-confined calcium indicator GCaMP6s over 350 × 280 × 120-µm3 FOV at 10 Hz.
For purposes of evaluation, the researchers compared the performance of HyLFM to the conventional technique of iterative light-field deconvolution (LFD), as well as to LFD improved by the technique of deep learning-based content aware restoration (LFD + CARE). The evaluation results demonstrated that HyLFM achieved superior overall performance on unseen data to conventional LFD and on par with the LFD + CARE combination both qualitatively and quantitatively, despite being dramatically faster.
The new approach exemplifies the growing role of big data solutions in the life sciences and related disciplines given the sheer quantities and amounts of data needed for analysis.
"Our method will be really key for people who want to study how brains compute," said co-author Robert Prevedel, PhD, group leader at the European Molecular Biology Laboratory in Heidelberg, Germany. "Our method can image an entire brain of a fish larva, in real time."
Do you have a unique perspective on your research related to microscopy? Contact the editor today to learn more.
Copyright © 2021 scienceboard.net