June 29, 2020 -- Strategically folded DNA arranged to mimic viral antigens -- known as DNA "origami" -- may produce strong immune responses, according to a June 29 report in Nature Nanotechnology. Researchers are now working on adapting this approach for the development of a vaccine against SARS-CoV-2.
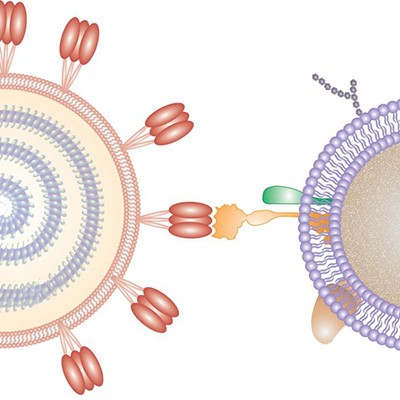
Virus-like particles displaying immunogens -- antigens that are specifically bound by components of the immune system that evoke an immune response -- can enhance B-cell activation and vaccine efficacy. However, the effects of antigen copy number, spacing, and affinity, as well as dimensionality and rigidity of scaffold presentation on B-cell activation, are poorly understood. Studies exploring the effect of antigen organization in this context have only allowed for limited variations in design.
DNA origami was invented in 2006 by Paul Rothemund of the California Institute of Technology. It involves the folding of DNA to create 2D and 3D shapes at the nanoscale. In 2016, a team of researchers from the Massachusetts Institute of Technology (MIT) developed an algorithm that can automatically design and build 3D virus-like shapes using DNA origami. This method offers a high degree of control to attach molecules at very specific locations.
"The DNA structure is like a pegboard where the antigens can be attached at any position," explained senior author Mark Bathe, PhD, MIT professor of biological engineering and an associate member of the Broad Institute of MIT and Harvard, in a statement. "These virus-like particles have now enabled us to reveal fundamental molecular principles of immune cell recognition for the first time."
Using DNA origami to study a clinically relevant antigen
The researchers sought to investigate the roles of immunogen using scaffolded DNA origami nanoparticles. To begin, they focused on HIV-1 envelope glycoprotein antigen gp120 (eOD-GT8). This antigen was specifically designed to bind with high affinity to a precursor of a CD4 binding site-specific HIV broadly neutralizing antibody (gIVRC01) to activate naïve B cells.
The team used DNA origami to spatially present the antigen at a variety of distances and densities. They created a 3D icosahedral nanoparticle with a 40-nanometer diameter size that is comparable to an HIV-1 envelope glycoprotein (60mer) and a 1D rigid-rod six-helix bundle around 80 nanometers long. By hybridizing the antigen to free single-stranded DNA overhang strands, the researchers were able to present antigen-varying spatial organizations.
While there has been a great deal of interest in using virus-like particle structures to drive optimal B-cell responses for vaccines, until now, the rules for how to design the display antigen have not been well understood, according to Irvine.
Best fit does not always indicate efficacy
Antigen conjugation and the ability of various designed nanoparticles to activate B-cell assays revealed that signaling is maximized by as few as five antigens. In fact, increasing copy numbers to 30 or 60 had no further effect on cellular responses. Moreover, the researchers determined that an interantigen distance of 25-30 nanometers resulted in increased B-cell receptor activation.
"It is often assumed that the higher the antigen density, the better, with the idea that bringing B cell receptors as close together as possible is what drives signaling. However, the experimental result, which was very clear, was that actually the closest possible spacing we could make was not the best. And, as you widen the distance between two antigens, signaling increased," said senior author Darrell Irvine, PhD, a member of the Ragon Institute and associate director at MIT's Koch Institute for Integrative Cancer Research.
The findings from the study have informed HIV vaccine design. The antigen investigated is now being tested in human clinical trials, using a protein nanostructure scaffold.
Interchangeable platform could lead to COVID-19 therapies
Recently, in response to the global COVID-19 pandemic, the team has created a variant of the HIV vaccine that swaps out the HIV antigens for a protein found on the surface of the SARS-CoV-2 virus. This work is being conducted with researchers from the Ragon Institute. The new vaccine is being tested in isolated B cells and animal models to determine if it can produce an effective immune response against the SARS-CoV-2 virus.
"Our platform technology allows you to easily swap out different subunit antigens and peptides from different types of viruses to test whether they may potentially be functional as vaccines," said Bathe.
If successful, this dynamic shift from HIV to SARS-CoV-2 would demonstrate the platform's ability to attach antigens from various viruses to the same DNA scaffold. Furthermore, it could be used to design variants that target multiple types of coronaviruses -- past and future -- for potential pancoronavirus vaccines.
Do you have a unique perspective on your research related to vaccine development or virology? Contact the editor today to learn more.
Copyright © 2020 scienceboard.net