March 6, 2020 -- A new technology called µMap uses photocatalysts, molecules that spur a chemical reaction when activated by light, to identify spatial relationships of cell surface proteins. The results are described by scientists from Princeton University and Merck in the March 6 edition of Science.
Cell surfaces have localized proteins that create microenvironments that play a crucial role in intercellular communication. Biologists have long sought improved methods to map microenvironments in the areas of proteomics, genomics, and neuroscience, just to name a few. Recent advancements in label targeting technologies have allowed for the specific labeling of proteins.
"For the technologies we have right now, the problem is not whether you can tag things," said study co-author David MacMillan, PhD, the James S. McDonnell Distinguished Professor of Chemistry at Princeton University, in a statement. "The problem is that you can tag thousands of things and so you can't tell what's way over there and what's right next door. That turns out to be really, really important because molecules or proteins or enzymes that signal each other are usually right next door to each other. Well, the state-of-the-art doesn't tell you what's close."
MacMillan and co-researchers decided to address the issue by leveraging the qualities of carbenes, which readily crosslink with biomolecules and cannot move farther than 4 nanometers (nm) away. They used these molecules to develop a new system in the form of diazirine-based probes that require excitation with ultraviolet light. They built on previous research indicating that light-powered iridium catalysts allow blue light-emitting diodes (LEDs) to indirectly activate diazirines, localizing the generation of carbenes within 0.1 nm of a photocatalyst (Science, March 6, 2020, Vol. 367:6482, pp. 1091-1097).
Called µMap, the system is an antibody-targeted photocatalytic diazirine activation that identifies neighbors around a particular protein within a radius of 1 to 10 nm. (For reference, a human hair is about 100,000 nm across.) Resolution on this level identifies the 10 or 15 closest molecules, offering sufficient spatiotemporal resolution to profile nanoscale protein assemblies on the surface of cells.
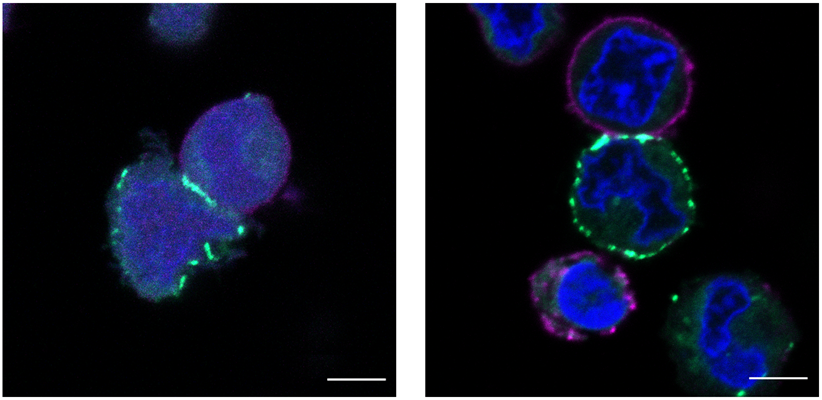
The researchers then applied µMap to the surface of two types of live cells. First, they selected CD45, a highly abundant tyrosine phosphatase on T-cell surfaces that is involved in antigen receptor signaling. The results showed specific binding of CD45 with two known associators (CD45AP and CD2). The µMap was also able to discriminate between the unrelated microenvironments of CD45, CD29, and CD47 on T cells.
Next, the µMap was applied to proximal protein interactome of programmed-death ligand 1 (PD-L1) in B cells. PD-L1 plays an important role in cancer cells as an immune checkpoint ligand that can accelerate tumor progression through suppression of T-cell activity. By identifying proteins that interact with PD-L1, researchers may be able to improve immunotherapies that target PD-L1 in cancers.
In the study, the µMap revealed that CD30 (a member of the tumor necrosis factor receptor family) and CD300A (an immune inhibitory receptor) interact with PD-L1, and it provided new insights into the microenvironments of checkpoint proteins.
"We've invented this tool that can give you a lot of information about these cancer cells," MacMillan said. "We think that by using this information, you can start to target those proteins as a way to also remove interfering signals. And if you can remove those signals, you make your immune system better at going after these cancer cells."
Do you have a unique perspective on your research related to molecular biology? Contact the editor today to learn more.
Copyright © 2020 scienceboard.net






