November 10, 2020 -- The likelihood of SARS-CoV-2 developing resistance to COVID-19 vaccines currently under development can be determined using repurposed blood and nasal test samples that are already being collected as part of clinical trials, according to Pennsylvania State University researchers. The perspective piece was published in PLOS Biology on November 9.
Vaccine resistance -- the increased ability of a pathogen to infect, replicate, and/or transmit from a vaccinated host -- is achieved through mechanisms such as serotype replacement, antigenic change, or increases in disease severity. Vaccine resistance occurs much less often than drug resistance.
For instance, the measles vaccine has been widely used for decades without the virus ever evolving the ability to transmit through vaccinated hosts. On the other hand, Streptococcus pneumoniae quickly evolved resistance to the pneumococcal conjugate vaccine (PCV7), and as a result, a new vaccine had to be developed (PCV13).
"A COVID-19 vaccine is urgently needed to save lives and help society return to its pre-pandemic normal," said co-author David Kennedy, PhD, assistant professor of biology at Penn State, in a statement. "As we have seen with other diseases, such as pneumonia, the evolution of resistance can quickly render vaccines ineffective. By learning from these previous challenges and by implementing this knowledge during vaccine design, we may be able to maximize the long-term impact of COVID-19 vaccines."
To date, most cases of vaccine resistance have been attributed to the absence of three factors:
- Induction of protective immune responses by targeting multiple virus epitopes simultaneously.
- Suppression of pathogen growth within hosts.
- Vaccine-induced immune responses that protect against all circulating serotypes.
According to the authors, when these vaccine features are missing, the probability of resistance increases, giving the virus multiple opportunities to mutate. Alternatively, when a vaccine contains all the listed features, the chances of resistance become miniscule.
As most of the vaccines under development specifically target the SARS-CoV-2 spike protein, if the virus were to evolve resistance against one vaccine, it could simultaneously undermine many others, otherwise known as "collateral" or "cross" resistance.
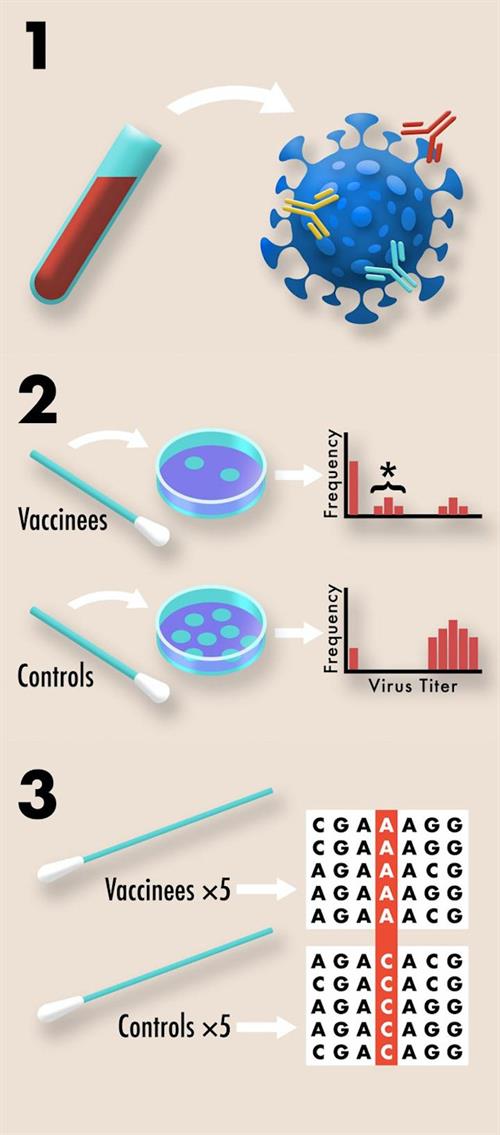
To avoid this scenario, the researchers suggested that test samples from clinical trials can be repurposed to assess the risk of resistance evolution, even prior to vaccine licensure.
Repurposing blood samples
Leveraging blood samples that are collected during all COVID-19 clinical trials to quantify antibody titer and serum neutralization, the samples can also be used to quantify redundancy of immune protection generated by various vaccine candidates.
"Much like how combination antibiotic therapy delays the evolution of antibiotic resistance, vaccines that are designed to induce a redundant immune response -- or one in which the immune system is encouraged to target multiple sites, called epitopes -- on the virus's surface, can delay the evolution of vaccine resistance," said co-author Andrew Read, director of the Huck Institutes of the Life Sciences at Pennsylvania State University. "That's because the virus would have to acquire several mutations, as opposed to just one, in order to survive the host immune system's attack."
Furthermore, diverse T-cell responses in vaccinated individuals can also delay resistance evolution. Quantifying redundancy of immune protection can therefore provide critical information for determining the likelihood of resistance evolution.
Repurposing nasal swabs
The researchers also proposed that nasal swabs collected from vaccinated and control individuals, normally used to quantify vaccine protection against infection, can also be used to acquire viral titer data as an easily collectible proxy for viral transmission. As suppressing pathogen transmission is key to preventing spread of disease, it is also important for preventing resistance. When a pathogen is not easily transmitted, this slows the evolution of resistance by minimizing opportunities for mutations to arise and reduces opportunities for natural selection to act on those mutations that do arise.
Nasopharyngeal swabs collected from symptomatic vaccinated and control individuals to confirm SARS-CoV-2 as the cause of disease can be repurposed to search for evidence of vaccine-driven selection by sequencing viral genomes of the samples. Tracking allele frequency between the viral genomes from vaccinated and control individuals can be used to identify natural selection towards resistance, according to the scientists.
"According to the World Health Organization, at least 198 COVID-19 vaccines are in the development pipeline, with 44 currently undergoing clinical evaluation," Kennedy said. "We suggest that the risk of resistance be used to prioritize investment among otherwise similarly promising vaccine candidates."
Do you have a unique perspective on your research related to immunology or virology? Contact the editor today to learn more.
Copyright © 2020 scienceboard.net