August 9, 2021 -- Researchers have demonstrated that prime editing -- a newer version of CRISPR-Cas9 gene editing -- can be successfully employed to correct gene mutations that cause cystic fibrosis. The new study, which tested the technique in human organoids, was published in Life Science Alliance on August 9.
Genome engineering has developed quickly since its inception more than a decade ago. Homology-directed repair uses an effector protein (Cas9) directed by guide RNA (gRNA) to a target site, which it uses to generate a double-stranded break. This strategy is easily reprogrammable to make gene edits; however, it is often prone to errors and can result in off-target editing.
To circumvent these issues, base editors were developed. Base editors contain a partially nuclease-inactive nickase-Cas9 (nCas9) protein that is fused to either an enzyme to convert cystine-guanine base pairs to thymine-adenine base pairs, or the reverse. These tools are very effective but are limited to DNA substitutions.
Thus, prime editing has been developed to insert or delete nucleotides without the need to generate double-stranded breaks. In this approach, Cas9 is fused to an engineered reverse transcriptase that is used to generate complementary DNA (cDNA) from an RNA template.
The fusion protein is combined with a prime editing guide RNA (pegRNA) that encodes a desired edit. Upon target recognition, the DNA is nicked, and the pegRNA becomes bound. The reverse transcriptase then synthesizes the edit and cellular DNA repair processes to resolve the opposing DNA strand to complete the edit.
Regarding the current study, researchers from University Medical Center Utrecht and Oncode Institute described how they use prime editing strategies to correct gene mutations that cause cystic fibrosis using human intestinal organoids.
"In our study, prime editing proves to be a safer technique than the conventional CRISPR-Cas9. It can build in a new piece of DNA without causing damage elsewhere in the DNA. That makes the technique promising for application in patients," said first author Maarten Geurts, a PhD candidate at the Hubrecht Institute, in a statement.
Proof of concept to correct cystic fibrosis mutations
Cystic fibrosis (CF) is a genetic disease that causes excessive mucus in the lungs, throat, and intestines, causing painful and deadly blockages. CF-causing mutations are localized in the cystic fibrosis transmembrane conductance regulatory (CFTR) channel, which is present in the cells of various organs, including the lungs. Due to the mutations, the channel does not function properly, leaving the layer of mucus that covers the cells with too little water. Consequently, the mucus becomes "sticky."
In the study, the researchers used intestinal organoids as an in vitro model of cystic fibrosis. The addition of a substance called forskolin causes healthy organoids to swell, but this does not happen in organoids with mutations in the CFTR channel. The team has previously shown that the response to forskolin can be used as a direct functional readout for repair of the CFTR gene in organoids derived from cystic fibrosis patients by base editing and CRISPR-mediated homology-dependent repair.
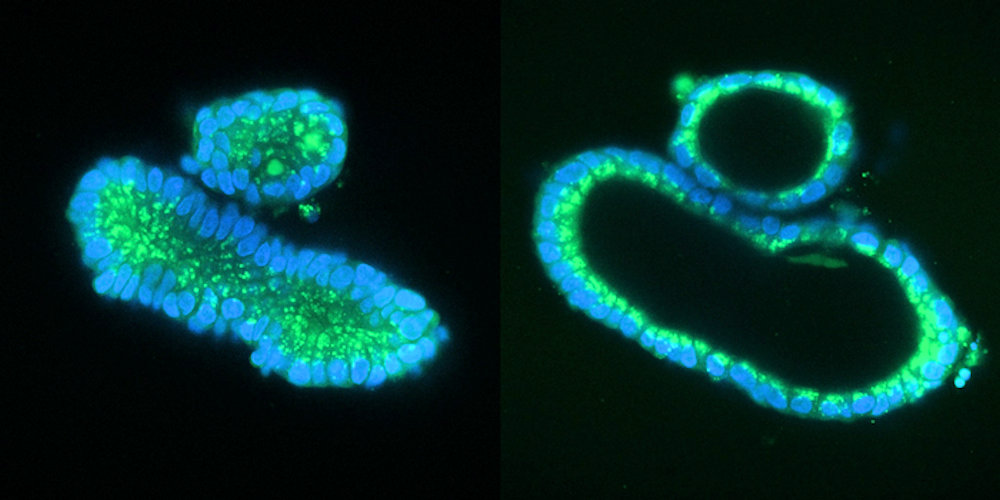
"We applied prime editing to the mutations, after which the treated organoids demonstrated the same response as the healthy organoids: They became swollen. That provided us with proof that our technique worked and replaced the mutated DNA," explained Geurts.
Using various iterations of desired targets with prime editing, the team attempted to repair the CFTR-F508del mutation. Even though efficiencies are low and undesired outcomes may occur, prime editing can repair the CFTR-F508del mutation in patient-derived intestinal organoids, the authors noted. When targeting the CFTR-R785 mutation, prime editing achieved complete repair of CFTR function in these organoids.
The authors pointed out the formation of unintended indels and undesired edits in some cases of prime editing. They suggest that this is explained by the need to generate a second nick on the opposing DNA strand, but that the use of two single-guide RNAs for the opposing strand could increase specificity of the system. There are still some challenges that lie ahead for the research team; for instance, the technique still needs to be adapted for safe use in humans.
"But this is a great step towards successfully applying prime editing in the clinic," said Geurts.
"New variants of CRISPR-Cas9, such as prime editing, can safely correct mutations without causing damage in other regions of the DNA," Geurts explained. "This will hopefully enable us to cure or even prevent genetic diseases in the future."
Do you have a unique perspective on your research related to gene editing or rare diseases? Contact the editor today to learn more.
Copyright © 2021 scienceboard.net









