February 17, 2022 -- The ability to sequence RNA in single cells has given scientists an unprecedented level of resolution in studying rare cell types. However, current approaches are designed to deal with a large number of cells, making it difficult to work with small samples. New research, published in Nature Methods on February 14, has described a technique using machine vision to detect cells and make single-cell RNA sequencing more efficient at a smaller scale.
Sequencing mRNA provides scientists with tremendous insight into genetic activity that drives the behavior of our cells. Thanks to advances in molecular biology and microfluidics over the last decade, it is now possible to sequence mRNA from individual cells in a high-throughput fashion. This level of detail means that even cells that are seemingly identical under the microscope can be distinguished based on their gene activity.
Single-cell sequencing has become popular with researchers seeking to find new cell types and understand the heterogeneity in tissues. Compared to sequencing bulk samples, the single-cell approach prevents the more common cells in a sample from overwhelming the signal of gene activity from rare cell types.
To support efforts to find rare cells or to build cell atlases in the body, the techniques for single-cell studies are designed to work with large samples. These methods either require a minimum input of thousands of cells or are simply not cost-effective for small quantities. This constraint could limit research in areas where only small samples are available, such as early-stage embryonic development.
Now, researchers led by Bart Deplancke, PhD, a professor of systems biology and genetics at the Swiss Federal Institute of Technology Lausanne in Switzerland, have developed a new approach that can cope with as few as 50 cells. The technique, called DisCo, uses machine vision to precisely control a microfluidic device so that cells are efficiently processed.
Single-cell RNA sequencing techniques typically rely on capturing an individual cell and an mRNA capture bead in a droplet. The bead is an essential part of the sequencing process -- the process does not work unless a bead is matched to precisely one cell. Various microfluidic techniques are designed to do this, but with a focus on throughput rather than efficiency.
With machine vision to detect the cells passing through the microfluidic device, DisCo was 75% efficient at capturing them correctly -- more than double the rate in many established platforms. The new system achieved this with a cost of only $100 for processing 100 cells, less than half the cost of other techniques for samples of this size.
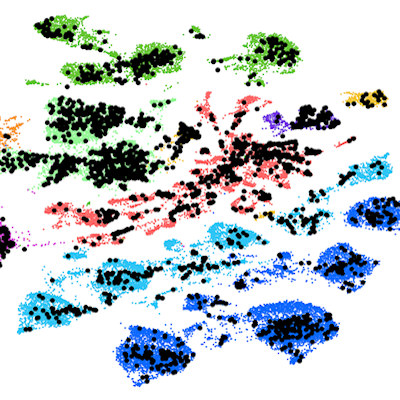
To test the system, the team analyzed organoids -- microscopic 3D tissues consisting of only a few dozen cells grown in a lab. These organoids can form strikingly different shapes and sizes when grown from single intestinal stem cells, despite being exposed to seemingly identical conditions.
With DisCo, individual organoids could be analyzed separately, which revealed that some had unusual cell type populations. Of particular interest, the researchers found one organoid subtype, which they called a "gobloid" that predominantly consisted of mucus-secreting goblet cells.
The authors of the paper wrote that they believe this technology will help engineer more robust organoid systems. Improving the realism of organoids could lead to advances in fields ranging from basic science to toxicology testing.
The system could also be useful in other domains where samples are limited, with the authors adding that they "expect this approach to be applicable to rare, small clinical samples to gain detailed insights into disease-related cellular heterogeneity and dynamics."
Do you have a unique perspective on your research related to single-cell technologies? Contact the editor today to learn more.
Copyright © 2022 scienceboard.net