February 22, 2021 -- Scientists have developed a new technique for determining alternative structural RNA shapes of the SARS-CoV-2 virus. These self-regulatory segments of RNA, called switches, could serve as potential antiviral drug targets, according to a communication published in Nature Methods on February 22.
The linear sequence of certain bacterial RNA genomes encodes all the proteins that are needed to take over the machinery of host cells. However, the RNA also folds back on itself, forming secondary and tertiary conformations that protect the bacteria from chemical modification or degradation.
Alternative ways of forming these secondary and tertiary structures will affect the way in which the RNA functions. For instance, riboswitches -- or regulatory segments that are found in certain RNA molecules -- either allow or block translation of associated genes into proteins. The folding of these RNA segments depends on the external environment.
Viral RNA also folds, and the switch between alternative conformations can be important to the life cycle of a virus.
"Whenever you see a dynamic structure in the RNA, this suggests a regulatory system," said Danny Incarnato, PhD, a molecular geneticist at the University of Groningen. "So, we devised a method to find in experimental data whether an RNA can form different conformations."
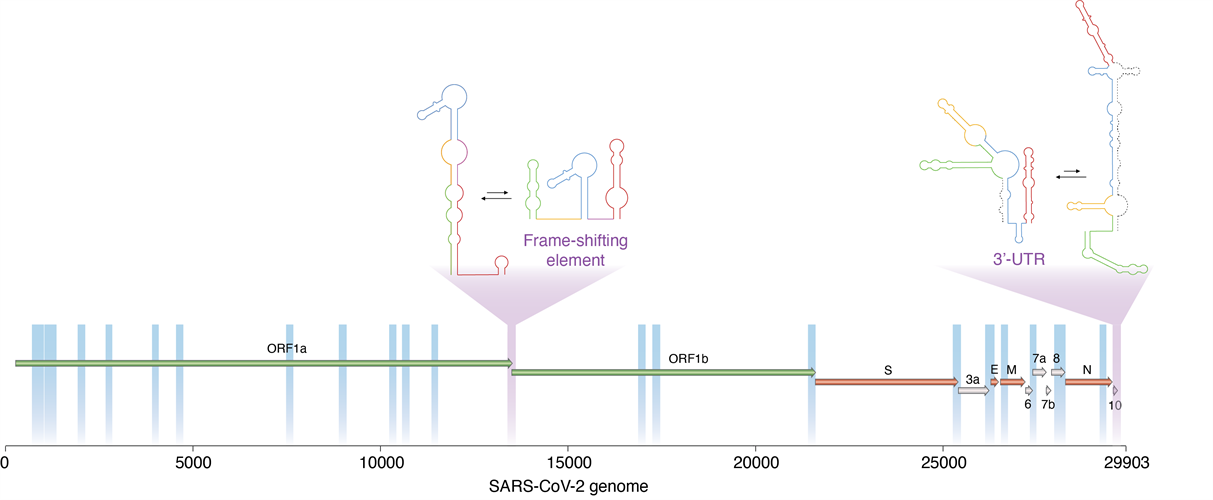
A new technique developed by researchers enabled them to determine which regions of the SARS-CoV-2 virus are dynamic, and thus able to switch between alternative, mutually exclusive conformations. These include two important viral regulatory regions (marked in purple). Image courtesy of Danny Incarnato, PhD, University of Groningen.
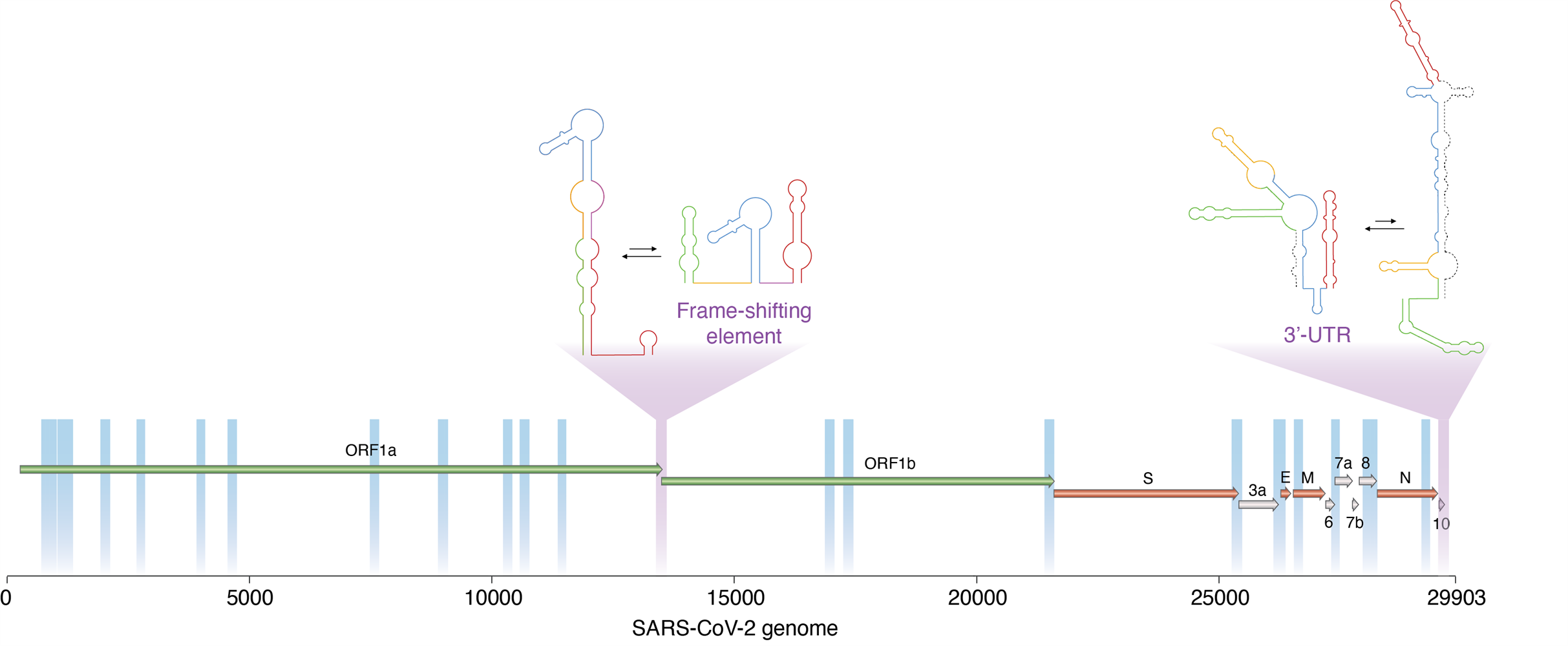
A new technique developed by researchers enabled them to determine which regions of the SARS-CoV-2 virus are dynamic, and thus able to switch between alternative, mutually exclusive conformations. These include two important viral regulatory regions (marked in purple). Image courtesy of Danny Incarnato, PhD, University of Groningen.
Past attempts at predicting alternative conformations of viral RNA structures have been limited and are based on the detection of unpaired ("mutated") nucleobases within the entire RNA sequence. These modified nucleobases are read as "mutations" during RNA transcription.
"The process is not very efficient and, therefore, not all the unpaired bases will be mutated," said Incarnato.
Further adding to the complexity of this process is that the same RNA segments can fold in multiple ways.
Inspection of RNA conformations
Researchers at the University of Groningen in collaboration with a team from the University of Torino have developed Draco, an algorithm for the deconvolution of coexisting RNA conformations from mutational profiling experiments, by sequencing and modeling 150-nucleotide-long fragments of RNA.
"The idea is to see which mutations occur together in a particular region and which do not," explained Incarnato. "This pattern points to alternative conformations. We created an algorithm that can rapidly analyze the huge number of reverse-transcribed RNA molecules."
The researchers applied the Draco algorithm to the analysis of the SARS-CoV-2 RNA genome structure. They found that the data-driven RNA structure predicted by Draco produced models that were nearly identical to reference structures and experimental data.
In the experiments, the Draco analysis identified 22 windows (accounting for around 15% of the SARS-CoV-2 genome) that folded consistently into two distinct conformations.
One enriched area of the genome spanned the open reading frame (ORF)1a-ORF1b boundary, overlapping with the frameshifting element, a conserved stem-loop of RNA that during translation can result in the production of multiple, unique proteins from a single RNA.
The data suggest that this region likely folds into either a single extended stem-loop structure or two stem-loop structures, rather than the traditional pseudoknotted structure, thus confirming that two conformations exist.
A second enriched area of the genome at the three prime untranslated region (3' UTR), the translation termination codon, also showed evidence of two conformations. The major conformation (63%) was phylogenetically similar to those observed in other sarbecoviruses. Alternatively, the minor conformation (36%) was predicted to form an alternative three-way junction, occluding both the bulged stem-loop (BSL) and distal stem-loop helices. The presence of these two conformations is supported by an independent analysis of RNA-RNA interactions in SARS-CoV-2 infected cells.
"It helps us to understand the `RNA structurome' and the effect it has on viruses and cells," said Incarnato.
The authors suggested that certain RNA mutations have been overlooked because the sequence encoded does not change, but rather the structure of the RNA molecule does. However, these RNA switches can still play a major role in diseases.
Do you have a unique perspective on your research related to infectious diseases or virology? Contact the editor today to learn more.
Copyright © 2021 scienceboard.net