June 30, 2020 -- To successfully infect human cells, SARS-CoV-2 may hijack host proteins in target cells to promote its own replication. Researchers may be able to leverage this information to identify and recommend drugs, according to a June 29 Cell article.
Viruses are unable to replicate and spread on their own, and instead must take control of a host cell's machinery and manipulate it to support viral replication and infection. Sometimes, this occurs by modifying host proteins, whereby chemical modifications can alter their structure and function.
One example of this process is phosphorylation -- the addition of a phosphoryl group to a protein by kinase enzymes. Kinases are key regulatory enzymes that play an essential role in many cell processes. Altering phosphorylation patterns of host proteins by viruses can potentially promote viral replication and transmission to other hosts.
According to the international team of researchers, proteomics approaches can offer a powerful way to quantify global changes in protein abundance and phosphorylation to elucidate mechanisms of viral pathogenesis, specifically by providing insight into how cellular pathways and processes are rewired upon infection.
The power of phosphoproteomics analysis for COVID-19
In the current study, the team used a mass spectrometry approach to study changes in protein abundance and phosphorylation during SARS-CoV-2 infection. Large changes in protein phosphorylation were observed within 24 hours of infection, indicating that the virus makes use of host post-translational regulatory systems to promote changes in cellular signaling. They found that 12% of host proteins that interact with the virus were modified.
The researchers suggest that kinases are hijacked during infection and most likely regulate the modifications of host proteins. Based on the phosphoproteomics survey, 97 of the 518 human kinases measured were changed by infection. Elevated p38 mitogen-activated protein kinase (MAPK) and casein kinase II (CK2) activation suggests that the virus induces changes in cytoskeleton organization of host cells.
The researchers observed that an increase in p38/MAPK signaling pathway activity occurred late in the time course, likely reflecting a more advanced stage of infection. The p38/MAPK pathway controls the production of potentially harmful pro-inflammatory cytokines, which is also observed in SARS-CoV-2 infection. However, the specific impact of p38/MAPK inhibition during SARS-CoV-2 remains unknown.
Alternatively, upregulated CK2 activation, in combination with reduced activity of cyclin dependent kinase (CDK1/2) activity during SARS-CoV-2 infection, led to a synthesis/gap 2 phase arrest, which may benefit the virus by ensuring its abundant supply of nucleotides and other essential host DNA proteins.
Learning more about SARS-CoV-2 mechanisms
"The virus prevents human cells from dividing, maintaining them at a particular point in the cell cycle. This provides the virus with a relatively stable and adequate environment to keep replicating," explained author Pedro Beltrao, PhD, group leader at European Molecular Biology Laboratory - European Bioinformatics Institute (EMBL-EBI), in a statement.
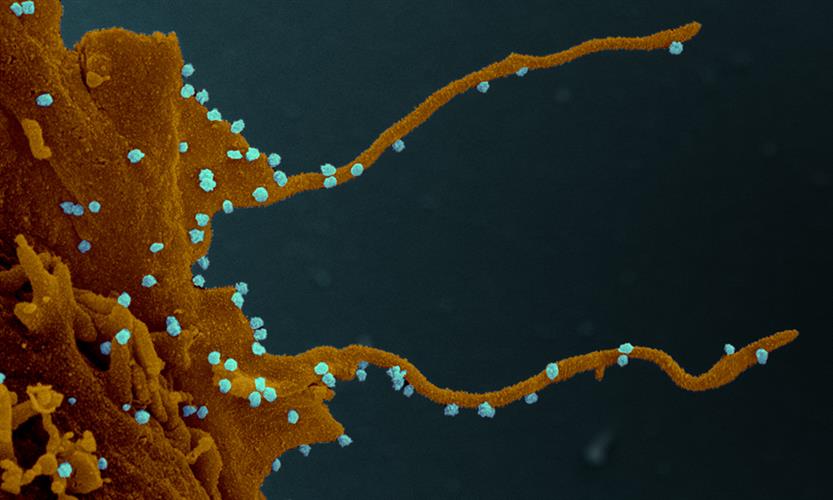
Increased phosphorylation of cytoskeleton protein targets and direct interaction with the actin assembly-inducing complex demonstrate the virus's ability to regulate the cytoskeleton during infection. Immunofluorescence imaging of infected cells reveals the formation of actin-rich filopodia containing viral proteins. It seems that SARS-CoV-2 protein clusters form along the length of filopodial protrusions and could be important for SARS-CoV-2 egress and/or cell-to-cell spread within epithelial monolayers.
"The distinct visualization of the extensive branching of the filopodia once again elucidates how understanding the biology of virus-host interaction can illuminate possible points of intervention in the disease," said author Nevan Krogan, PhD, director of the University of California, San Francisco Quantitative Biosciences Institute (UCSF QBI) and senior investigator at Gladstone Institutes.
Drug repurposing with kinases in mind
Subsequently, based on results observed from the phosphoproteomics analysis, bioinformatics can be employed to identify regulated kinases from phosphorylation profiles and help identify potential SARS-CoV-2 therapies. The team found antiviral activity for several drugs and compounds that are either U.S. Food and Drug Administration approved, in clinical testing, or under preclinical development.
These include silmitasertib (CK2 inhibitor in phase II), ralimetinib (p38 inhibitor in phase II), ARRY-797 (p38 inhibitor in phase II/III), and other kinase blockers that represent potential COVID-19 therapies. The researchers note the specific use of kinase inhibitors in combination with other drugs for the treatment of SARS-CoV-2, noting remdesivir as a suggested combinatorial agent.
"Kinases possess certain structural features that make them good drug targets. Drugs have already been developed to target some of the kinases we identified, so we urge clinical researchers to test the antiviral effects of these drugs in their trials," Beltrao said.
The researchers plan to build upon their work by testing additional kinase inhibitors while identifying underlying pathways and additional therapeutic targets against SARS-CoV-2.
Zoic Labs has partnered with researchers at the UCSF QBI to develop an interactive data visualization tool. It allows researchers to share their findings and characterizes critical connections between the 27 SARS-CoV-2 proteins and the 332 human genes they interact with.
The research collaboration included researchers from EMBL-EBI, the QBI Coronavirus Research Group in the School of Pharmacy at UCSF, the Howard Hughes Medical Institute, the Institut Pasteur, and the Centre for Integrative Biological Signalling Studies of the University of Freiburg.
Do you have a unique perspective on your research related to bioinformatics or cell biology? Contact the editor today to learn more.
Copyright © 2020 scienceboard.net