February 12, 2020 -- An inventory of over 350 DNA sequences from phage genomes revealed that these genomes can be over 200,000 base pairs (bp) in length, with the largest phage genome at 735,000 bp in length. The report published in Nature on February 12 evaluated the prevalence, diversity, and ecosystem distribution of phages with large genomes.
Bacteriophages (phages) are bacteria-killing viruses that contain genes normally found in bacteria, which they use against their bacterial hosts in a virus-like manner. The median genome size for phages is around 52,000 bp. Before this study, phages with genomes of more than 200,000 bp were referred to as "jumboaphages" and phages with genomes of more than 500,000 bp were called "megaphages," but their occurrence was thought to be quite rare.
Diversity of huge phages
In the current study, researchers from the University of California (UC), Berkeley demonstrated that large-phage genomes, which they call "huge phages," are more common than previously described. By analyzing data from many databases of DNA, they found large-phage genomes originated from nearly 30 unique Earth environments ranging from infant guts and pregnant women to Tibetan hot springs, a South African bioreactor, hospital rooms, oceans, lakes, and underground.
"We are exploring Earth's microbiomes, and sometimes unexpected things turn up. These viruses of bacteria are a part of biology, of replicating entities, that we know very little about," said Jill Banfield, PhD, a UC Berkeley professor of earth and planetary science and of environmental science, policy, and management, in a statement. "These huge phages bridge the gap between nonliving bacteriophages, on the one hand, and bacteria and Archaea. There definitely seem to be successful strategies of existence that are hybrids between what we think of as traditional viruses and traditional living organisms."
The researchers conducted phylogenetic analysis based on large terminase subunit proteins and major capsid proteins. Ten clades of huge phages were identified and named after words for "big" in the languages of the paper's co-authors: Mahaphage (Sanskrit); Kabirphage, Dakhmphage, and Jabbarphage (Arabic); Kyodaiphage (Japanese); Biggiephage (Australian); Whopperphage (American); Judaphage (Chinese); Enormephage (French); and Kaempephage (Danish).
The fact that these phages consistently grouped together within clades indicates that large genome size is a relatively stable trait. Moreover, phages that grouped together within a clade are likely to have replicated in bacteria of the same phylum. For example, in the study, genomes over 634,000 bp in length were all predicted to replicate in Bacteriodetes and cluster in the Mahaphage clade.
"Phages with large genomes are quite prominent across Earth's ecosystems, they are not a peculiarity of one ecosystem," Banfield said. "And phages which have large genomes are related, which means that these are established lineages with a long history of large genome size. Having large genomes is one successful strategy for existence, and a strategy we know very little about."
What's in huge phage genomes
Much of the DNA contained within these huge phages is part of the CRISPR-Cas systems, including type II, the recently described type V, new variants of the type V-U systems, and new subtypes of the type V-F system. One of the huge phages also produced Casø (ø has traditionally been used to denote bacteriophage), which is analogous to the Cas9 protein that is part of the revolutionary CRISPR-Cas9 gene-editing tool. It is likely that phages augment the CRISPR systems of host bacteria, most likely to target other viruses and prevent competition.
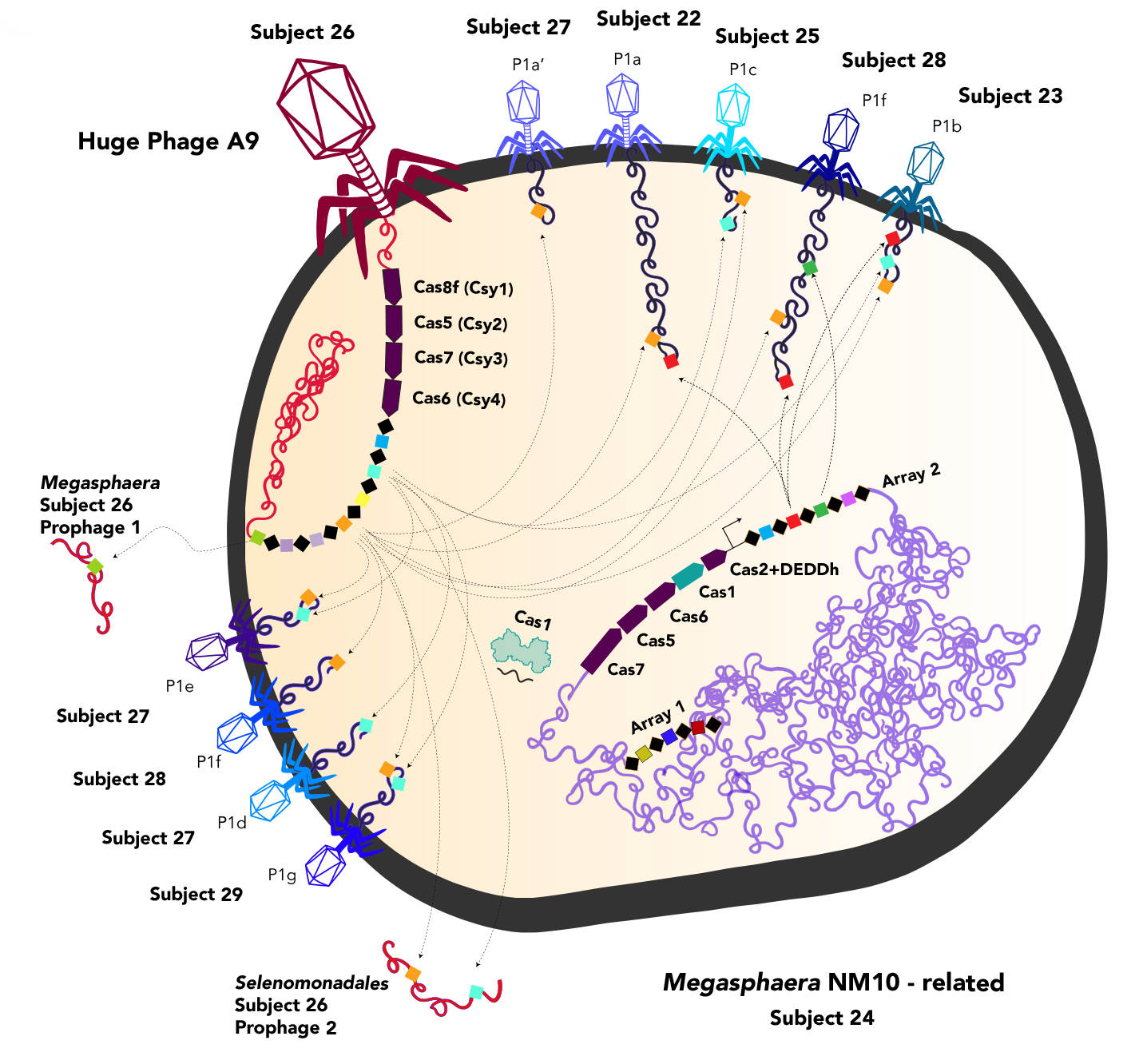
Understanding how phages interact with bacteria and viruses can help researchers elucidate mechanisms that confer antibiotic resistance and pathogenesis. Phages may play a key role in these processes, as they carry damaging genes into bacteria that colonize within humans.
"Some diseases are caused indirectly by phages, because phages move around genes involved in pathogenesis and antibiotic resistance," Banfield said. "And the larger the genome, the larger the capacity you have to move around those sorts of genes, and the higher the probability that you will be able to deliver undesirable genes to bacteria in human microbiomes."
The huge phages also contained genes that code ribosomes (transcription) and transfer RNAs (translation), which the researchers believe they use to make more copies of their own proteins at the expense of bacterial proteins.
"Typically, what separates life from nonlife is to have ribosomes and the ability to do translation; that is one of the major defining features that separate viruses and bacteria, non-life and life," said study co-author Rohan Sachdeva. "Some large phages have a lot of this translational machinery, so they are blurring the line a bit."
Do you have a unique perspective on your research related to genomics or cell biology? Contact the editor today to learn more.
Copyright © 2020 scienceboard.net






