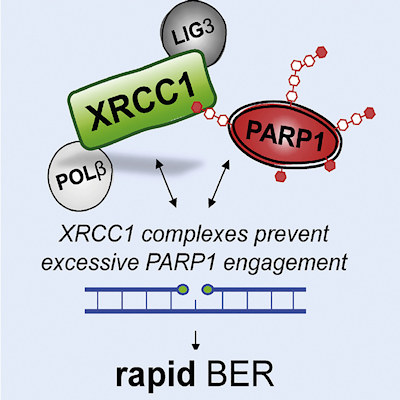
January 4, 2022 -- Researchers can now visualize DNA repair in unprecedented detail using a new high-throughput microscopy technique in combination with machine learning. The methodology, published in Cell Reports on December 28, has led to the identification of new proteins involved in DNA repair.
The human body suffers more than 10,000 DNA lesions every day. Accurate repair of DNA lesions is essential to prevent mutations and chromosomal rearrangements that could compromise an organism's survival and development. These repairs are facilitated by DNA damage response (DDR) factors. Quickly following a DNA lesion, DDR factors rapidly bind to damaged DNA to recruit specialized proteins that can then repair the DNA. If the repair mechanisms fail, it may lead to pathologies such as cancer, immunodeficiencies, neurodegeneration, and premature aging.
Identifying DDR factors has traditionally been achieved using loss-of-function screens. However, this methodology is limited by low throughput. Overexpression-based or gain-of-function screens, though, may allow scientists to further identify and study DDR factors.
In the study, researchers used a gain-of-function approach based on the expression of chromatin factors to screen for previously unknown modulators of DNA repair using two high-throughput, imaging-based DNA repair platform systems.
The first high-throughput microscopy assay provided a specifically tailored immunofluorescence readout of the kinetics of DNA repair based on the sequential accumulation and decrease of gamma-H2A.X variant histone (H2AX) and p53-binding protein 1 (53BP1) at breaks analyzed in a 384-well format. Using this system, the researchers detected the presence or absence of select DDR at DNA breaks. The researchers then applied machine-learning analysis to reveal chromatin factors that influence DNA repair over time.
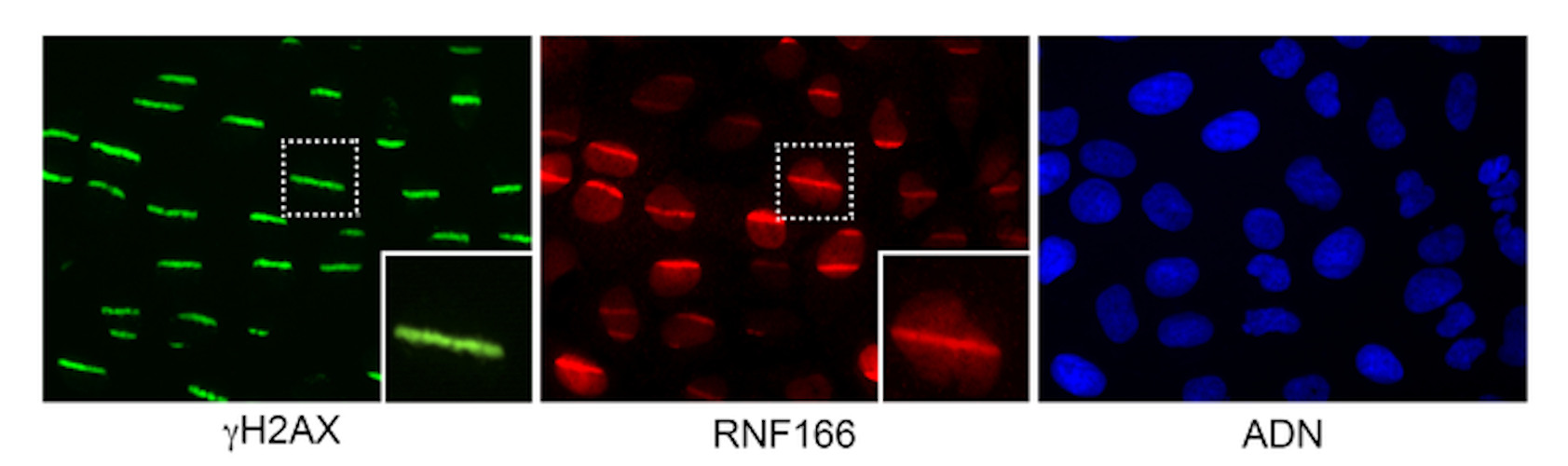
Next, the team built a complementary platform developed specifically to monitor the recruitment of tagged proteins to DNA breaks using gain of function of open reading frames. The second imaging screen determined the localization of more than 300 proteins to damaged chromatin in a single, rapid assay using high-throughput laser microirradiation. This assay allowed the researchers to visually monitor the proteins after DNA damage.
"We saw that many proteins adhered to damaged DNA, and others did just the opposite: they moved away from the DNA lesions," the authors noted. "The fact that they either bind to or remove themselves from damaged DNA, to allow the recruitment of repair proteins to the lesion, is a common feature of DNA repair proteins. Both phenomena are relevant."
These new screens have led to the identification of several candidates that, until now, have not previously been linked to DNA damage; that is, a protein that is recruited to or excluded from DNA breaks. The authors also provided proof-of-principle characterization of the plant homeodomain finger protein 20 (PHF20), which they discovered was excluded from DNA breaks within seconds following damage and was independent of poly (ADP-ribose) polymerase activity.
The study, the authors said, demonstrates that PHF20 competes with 53BP1 for binding to H4K20me2, inhibiting the recruitment of 53BP1 to double-stranded breaks and altering proper DNA repair. The new finding defines a system that should allow for the interrogation of novel factors modulating DNA repair in an unbiased way and in a high-throughput manner.
"Until now, one limiting factor in tracking DNA repair kinetics was the inability to process and analyze the amount of data generated from images taken by the microscope," said the authors.
The technologies offer new opportunities to study DNA repair and to manipulate it. An advantage is that both platforms are very versatile and can be used to discover new genes or chemical compounds that affect DNA repair.
"By knowing how DNA lesions occur and how they are repaired, we will learn more about how cancer develops and how we can fight it," said Bárbara Martínez, PhD, a corresponding author and a member of the metabolism and cell signaling group at the National Cancer Research Center in Madrid. "Any new discovery in DNA repair will help develop better cancer therapies, whilst protecting our healthy cells."
Do you have a unique perspective on your research related to cell biology or bioimaging? Contact the editor today to learn more.
Copyright © 2022 scienceboard.net