August 23, 2021 -- A new multidimensional dataset for COVID-19 multiomics research has been released to the public, along with a free online research portal for analyzing the data. The COVIDome Explorer and dataset, described recently in Cell Reports, give researchers a way to share data and collaborate in real-time for the clinical management of COVID-19.
Since the pandemic's start, research on SARS-CoV-2 has been fast and furious, and scientists have made significant progress in characterizing its virology and epidemiology. However, until now there has been no widely accessible, analysis-ready public dataset allowing the research community to generate and test hypotheses and to make real-time discoveries on the diverse molecular, cellular, and pathophysiological processes associated with COVID-19 in patients.
The newly released multiomics dataset, called COVIDome, includes data for 105 people, comprising 73 hospitalized COVID-19 patients and 32 COVID-19-negative controls. All COVID-19-positive participants were hospitalized due to moderate symptoms, but none had developed severe clinical disease requiring admission to the intensive care unit.
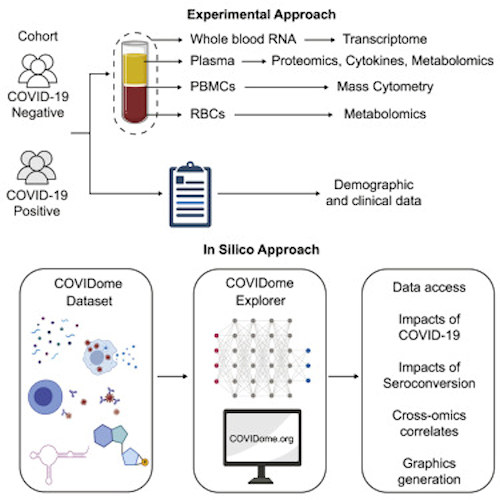
In addition to demographic and clinical data, the COVIDome dataset contains full multiomics analyses of the patients, including matched analysis of the whole-blood transcriptome using RNA sequencing, plasma proteomics, mass spectrometry and cytokine profiling, plasma and red blood cell metabolomics, deep immune cell phenotyping by mass cytometry, and seroconversion assays.
To facilitate use of the dataset, the same research team that assembled COVIDome simultaneously released COVIDome Explorer, an online researcher portal where the COVIDome data can be analyzed and visualized in real-time. The portal features six independent dashboards: Cohort (which includes deidentified demographic and clinical data), Transcriptome, Proteome, Cytokines, Metabolome, and Immune Maps.
The dataset and analysis tools were developed by scientists from the University of Colorado (CU) Anschutz Medical Campus, led by Joaquin Espinosa, PhD, a professor at the CU School of Medicine.
"Our mission for the COVIDome Explorer is to enable the development of better prevention, diagnostic and therapeutic tools for the clinical management of COVID-19," said Espinosa in a statement. "Although great progress has been made in all these areas, the speed of research for COVID-19 has been hampered by the lack of widely accessible, public datasets that can be analyzed and reanalyzed in real time by anyone."
In the paper, published on July 27, the authors demonstrated the ability of the COVIDome dataset, when analyzed using COVIDome Explorer, to reveal potentially biologically meaningful relationships among features across diverse omics datasets.
For example, using COVIDome, they were able to confirm that plasmablasts, which proliferate in COVID-19-positive individuals, are positively correlated with many immunoglobulin genes in the whole blood transcriptome, including IGKV4-1, IGHV6-1, and IGLC2.
In another example, the authors used COVIDome to identify the multiomics biosignatures associated with varying levels of C-reactive protein (CRP), an acute phase protein associated with increased probability of developing severe COVID-19. They were able to confirm positive correlations between CRP levels and the levels of serum amyloid proteins serum amyloid A1 (SAA1) and SAA2, the acute phase protein lipopolysaccharide binding protein (LBP), and the cytokines interleukin 6 (IL-6) and IL-10.
The authors encouraged users of COVIDome Explorer to find their own such "cross-omics correlations."
"We expect that this online portal will rapidly accelerate COVID-19 data sharing, hypothesis testing and discoveries worldwide," Espinosa said.
Quick and easy access to COVID-19 data is critical with cases on the rise due to the delta variant, said study co-author Dr. Thomas Flaig, vice chancellor of research at the CU Anschutz Medical Campus, in a statement.
"The global health crisis imposed by the COVID-19 pandemic further emphasizes the need for rapid collaboration, open access to manuscripts under review and data sharing," said Flaig. "Now more than ever, with the rise of the COVID-19 Delta variant and breakthrough infection rates rising, it's critical to have easy and timely access to COVID data to combat the pandemic and advance the best scientific data available."
The Explorer software was developed using the tools R, R Studio, and the R-based web application framework Shiny. The online portal is hosted on Shiny's cloud-based platform-as-a-service (PaaS) environment.
COVIDome Explorer can be accessed online at the CU Anschutz Medical Campus' website.
Do you have a unique perspective on your research related to bioinformatics? Contact the editor today to learn more.
Copyright © 2021 scienceboard.net