June 7, 2021 -- Targeting regions of the influenza virus that do not often change may be an effective strategy for developing next-generation universal flu vaccines, according to new research published in Science Translational Medicine on June 2. Researchers interrogated immune responses from the 2009 H1N1 influenza pandemic to determine which targets to include in new vaccines.
Hemagglutinin (HA) is the major surface antigen of influenza viruses and is composed of two major domains: the head and the stalk. Flu viruses evolve rapidly from season to season, leaving public health researchers to anticipate the predominant variation of the virus that will circulate each year. Seasonal influenza virus vaccines largely induce potently neutralizing antibodies against the immunodominant variable epitopes of the head domain, which provide narrow protection against only a few strains.
To further complicate the matter, sometimes new, unexpected variants emerge, resulting in seasonal vaccines that are not very effective. Therefore, the ultimate goal of influenza vaccine researchers is to develop a universal vaccine that can account for any virus strain or variation in a given year or even longer.
The new study evaluated immune responses of people who were first exposed to the 2009 H1N1 pandemic flu virus, either from infection or a vaccine. The team, led by University of Chicago immunologists Jenna Guthmiller, PhD, and Patrick Wilson, PhD, along with structural biologists Julianna Han, PhD, and Andrew Ward, PhD, from Scripps Research Institute, aimed at identifying a strategy for universal flu vaccines that targets conserved regions of the HA head domain.
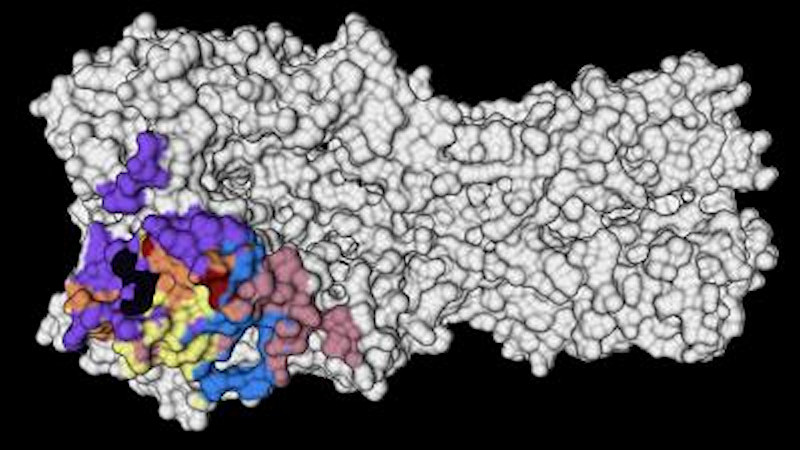
The head domain of HA is more variable and mutates more frequently than the stalk domain. There are five major antigenic sites of the HA head that exhibit strong overlap with the variable regions of the HA head. However, two epitopes, the receptor-binding site and the lateral patch, contain more conserved regions of the HA head. For this reason, the team focused its analysis on monoclonal antibodies (mAbs) targeting these two regions.
By examining antibody responses of individuals, the team determined that the first exposure to the 2009 pandemic H1N1 virus preferentially induces antibodies against conserved epitopes of the HA head. The virus has mutated since 2009, and the researchers found that in the absence of preexisting immunity against variable epitopes of the HA head of the virus, individuals can recall memory B cells targeting conserved epitopes of the HA head. Alternatively, repeated exposure to the virus elicits memory B cells against the variable major antigenic sites of the HA head.
Importantly, the team also determined that first exposure to the virus recalls memory B cells that are specific to the conserved receptor-binding site or lateral patch epitopes of the HA head domain. They suggested that antibodies elicited against those epitopes are broadly neutralizing against antigenically drifted and shifted H1N1 viruses and even against a recent H1N1 strain that has mutated near the lateral patch.
Previous research has shown that receptor-binding site antibodies exhibit broad cross-reactivity among H1N1 viruses and can occasionally cross-react with H3N2 viruses. The current study found that receptor-binding site mAbs used a diverse range of V(D)J genes across both the heavy and light chains.
Conversely, very little is known about lateral patch-binding antibodies, including their relative abundance, repertoire, and binding features. The data revealed that B cells targeting the lateral patch use a highly restricted heavy chain (VH) and variable region (VK) gene repertoire.
The researchers found that both receptor-binding site and lateral patch antibodies were broadly neutralizing. In fact, lateral patch-binding mAbs were able to bind 100% of 1918 H1 and 1977-2009 H1N1 viruses tested. Receptor-binding site antibodies cross-reacted with H3 and influenza B viruses.
This suggests that the key residues involved in monoclonal antibody binding to the H1 receptor-binding site are similar to those residues within the H3 and influenza B virus receptor-binding site epitopes.
The new study reveals more details about the conditions that can recall the same strong immune responses as this first exposure.
"That's the exciting thing about this study," Guthmiller said. "Not only have we found these broadly neutralizing antibodies, but now we know of a way to actually induce them."
The only problem is that on subsequent encounters with the virus or a vaccine, the body does not generate those same highly effective antibodies. Instead, the immune system tends to target newer variations on the virus. That may be effective at the time but is not helpful down the road when another, slightly different version of influenza comes along.
"When people encounter that virus a second or third time, their antibody response is pretty much completely dominated by antibodies against those more variable parts of the virus," Guthmiller said. "So that's the uphill battle that we continue to face with this."
To avoid this complication, the authors recommend that a combined epitope approach is necessary to generate a robust vaccine that is capable of protecting against viral resistance. They continue on to describe several platform technologies that can be modified to drive antibodies specifically to the receptor-binding site and lateral patch epitopes that could provide a framework for next-generation influenza vaccines that target the HA head and produce protective, broadly neutralizing antibodies.
In the past century, two of the four flu pandemics have been caused by H1N1 influenza, including the 1918 Spanish flu pandemic that killed as many as 100 million people. The findings of the new study are reassuring in the fight against possible future pandemics caused by other H1 viruses.
"The odds of there being another pandemic within our lifetime caused by an H1 virus is quite high," Guthmiller concluded. "Just knowing that we actually have the immune toolkit ready to protect ourselves is encouraging. Now it's just a matter of getting the right vaccine to do that."
Do you have a unique perspective on your research related to infectious diseases or virology? Contact the editor today to learn more.
Copyright © 2021 scienceboard.net