March 25, 2020 -- Scientists are urgently working to build the first complete all-atom model of the SARS-CoV-2 coronavirus envelope, estimated to contain over 200 million atoms. This work was conducted as a collaboration between the Amaro Lab of the University of California (UC), San Diego and the National Science Foundation-funded Frontera supercomputer at the University of Texas (UT) at Austin.
Rommie Amaro, PhD, professor of chemistry and biochemistry at UC San Diego, is leading efforts to understand molecular recognition. Most recently, the Amaro Lab used the Frontera supercomputer on March 12-13 to run nanoscale molecular dynamics simulations for the novel coronavirus on up to 4,000 nodes, or about 250,000 of Frontera's processing cores.
This work comes in the wake of the Amaro Lab's recent publication in ACS Central Science on the all-atom simulation of the influenza virus envelope. The work with the two viruses has a number of similarities, according to Amaro.
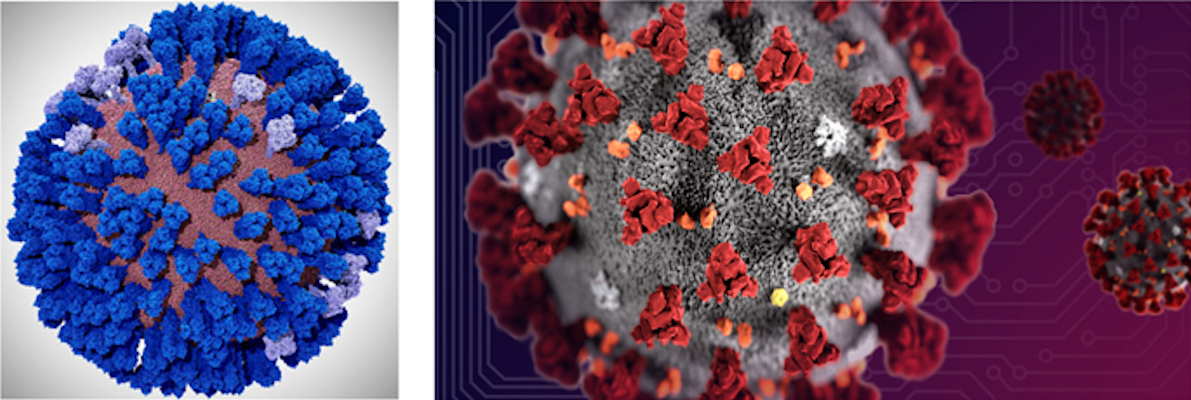
The simulations will provide insights into the infectivity of SARS-CoV-2. This information will help scientists on the front lines design new drugs and understand how current drugs and new potential drug combinations work.
"Hopefully the public will understand that there are many different components and facets of science to push forward to understand this virus," said Amaro. "These simulations on Frontera are just one of those components, but hopefully an important and a gainful one."
Copyright © 2020 scienceboard.net






