March 19, 2021 -- Bioengineers have demonstrated that simultaneous control over transcription and translation in cells can achieve the most stringent control of gene expression to date. The findings, published in Nature Communications on March 19, will open new avenues for improved biotechnologies and synthetic biology applications.
The ability to dynamically control gene expression has revolutionized biotechnology. Control of gene expression underpins a huge variety of applications in bioengineering.
Yet, the seemingly simple task of turning expression of a gene "on" or "off" is actually difficult to achieve because genes rarely have such discrete states or ever become completely silenced, leading to "leaky expression," which can wreak havoc on engineered systems.
In the past decade, synthetic biologists have developed advanced methods for controlling gene expression. These include engineered regulation processes based on DNA binding proteins (transcription activator-like effector nuclease [TALEN] and CRISPR interference [CRISPRi]), RNA-RNA interactions, post-trancriptional and post-translational processes (protein degradation), as well as directed evolution. These tools help researchers more strictly control gene expression that affects both transcription and translation with the goal of reducing unwanted expression.
However, multilevel regulation (i.e., independent and simultaneous control of transcription and translation) has not been achieved or implemented. Scientists from the University of Bristol studied the combined use of transcriptional and translational regulators to stringently control protein expression.
The team took inspiration from nature, where key events are often controlled by multiple processes simultaneously.
"If you look at a Venus flytrap you find that a trap will only close when multiple hairs are triggered together," said lead author Veronica Greco, PhD student at Bristol's School of Biological Sciences, in a statement. "This helps reduce the chance of a trap closing by accident. We wanted to do something similar when controlling the expression of a gene inside a cell, adding multiple levels of regulation to ensure it only comes on precisely when we want it to."
They designed, built, and tested a variety of synthetic multilevel controllers (MLCs) and compared the relative performance of each. The MLCs consisted of a gene encoding a transcriptional regulator and a second gene encoding a translational regulator. In the system, both genes are necessary for production of the gene of interest.
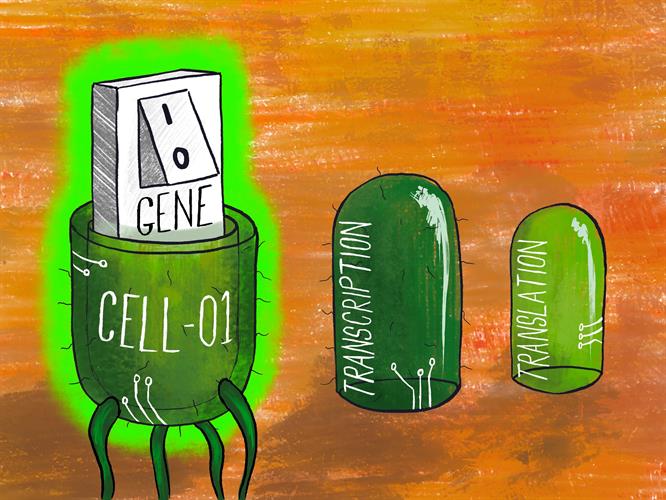
First, the controllers were implemented in a coherent type 1 feed-forward loop (C1-FFL) regulatory motif -- a regulatory path commonly found in Escherichia coli and yeast. In comparison to single-level controllers, MLCs were shown to suppress protein production by eliminating intrinsic promoter noise (variations in gene expression). This demonstrates the ability of MLCs to strictly control expression within cells.
"What was wonderful about this project was how well it worked to harness two of the core processes present in every cell and underpinning all of life - transcription and translation," added author Claire Grierson, PhD, head of the biological sciences school at Bristol.
Next, the team developed an eight-part genetic template and toolkit of parts that allowed for rapid combinatorial assembly of MLCs. The design enables both single- and multilevel regulation and has the option to introduce protein tags for further post-translational control of the gene of interest (e.g., through protein degradation). The toolkit contains eight types of part plasmids, and the final construct is inserted into a backbone plasmid.
The MLC designs were combined with three different RNA-based translational regulators including a toehold switch, a small transcription-activating RNA, and a dual-control system.
Escherichia coli cells were transformed with the various controller constructs, and green fluorescent protein (GFP) fluorescence was measured using flow cytometry to characterize the performance of each. The researchers found that inducible expression systems can be created with greatly reduced leaky expression when in an "off" state, while also maintaining high expression rates once induced.
Moreover, working in collaboration with researchers from the Max Planck Institute for Terrestrial Microbiology, the team was able to go a step further, showing that the synthetic genetic circuits using the MLCs could be used for cell-free protein synthesis in bioproduction platforms. The multilevel systems offered greatly improved control of gene expression, even in cell-free environments.
"When we engineer microbes, we often try to simplify our systems as much as possible, thinking we'll have better control over what is happening," said senior author Thomas Gorochowski, PhD, a Royal Society University Research Fellow at Bristol. "But what we've shown is that embracing some of the inherent complexity of biology might be the key to fully unlocking its potential for the high-precision biotechnologies of tomorrow."
Do you have a unique perspective on your research related to synthetic biology? Contact the editor today to learn more.
Copyright © 2021 scienceboard.net