September 26, 2019 -- It is now possible to rapidly image synaptic proteins at high-resolution thanks to a new technique developed by researchers at MIT and the Broad Institute of Harvard. Details of the technique which use fluorescent nucleic acid probes to label different proteins are described the September 26 Nature Communications article.
Neuronal synapses are fundamental sites of electrochemical signal transmission in the brain composed of hundreds of proteins that are responsible for development and function. Dysfunctional of these proteins can lead to conditions such as schizophrenia and autism. Characterizing these proteins is an important goal in both basic and translational neuroscience.
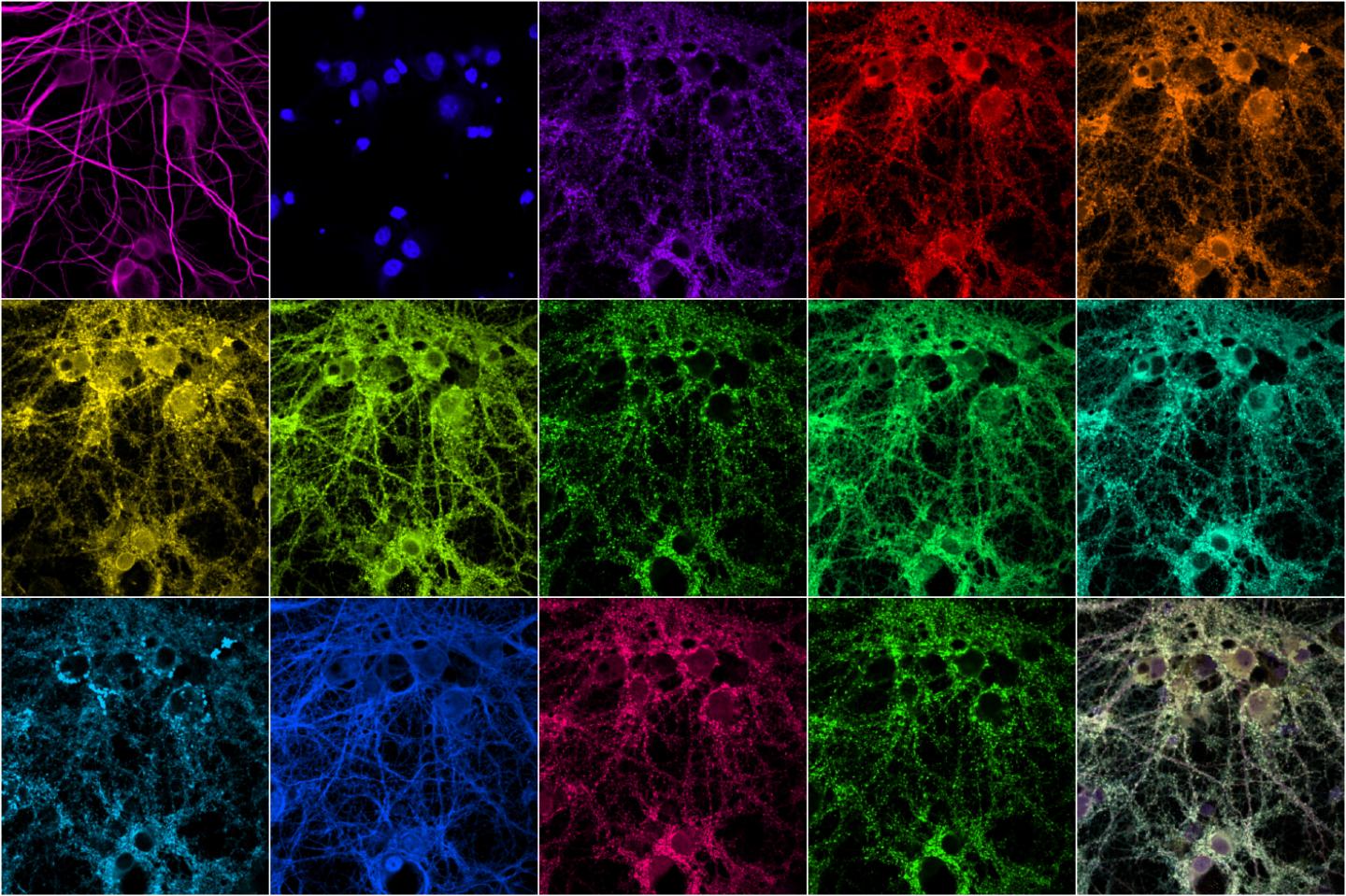
The research teams used multiplexed protein imaging to overcome the limitations of conventional fluorescence microscopy including a restricted number of proteins observed, and laborious methodologies. In the new method, the researchers used diffusible fluorescent imaging probes to target specific protein markers or antibodies. This allows for simultaneous antibody loading prior to imaging, rapid probe-exchange with mild buffers, and super-resolution imaging using Points Accumulation In Nanoscale Topography (PAINT). The DNA strands have an inherently low affinity for each other, so they bind and unbind periodically, creating a blinking fluorescence can be imaged using super-resolution microscopy. This technique is time-consuming, making it impractical for imaging many proteins in a large sample.
Therefore, the team used Probe-based Imaging for Sequential Multiplexing (PRISM) which fluorescently labels single-stranded DNA for imaging. The DNA-antibody complex binds tightly, locking nucleic acids into place. This provides high affinity and specificity with reduced background fluorescence from unused probes. The technique offers quantitative neuronal phenotyping using confocal imaging that was previously impossible using single-stranded DNA probes alone. Compared to PAINT, PRISM offers much faster experimental cycles.
"Multiplexed imaging is important because there's so much variability between synapses and cells, even within the same brain," says Mark Bathe, an MIT associate professor of biological engineering. "You really need to look simultaneously at proteins in the sample to understand what subpopulations of different synapses look like, discover new types of synapses, and understand how genetic variations impact them."
This technique was applied to confocal imaging of seven synaptic proteins imaged in neuronal culture simultaneously with five cytoskeletal proteins that interact with each other. The resultant multiplexed imaging data allowed form quantitative analysis of 66 protein co-expression profiles from thousands of individual synapses within the same neurons. One of the proteins they looked at is shank3, a scaffold protein that has been linked to both autism and schizophrenia. The researchers are currently investigating how the structure and composition of synapses are affected by knocking out genes associated with various disorders.
"Understanding how genetic variation impacts neurons' development in the brain, and their synaptic structure and function, is a huge challenge in neuroscience and in understanding how these diseases arise," Bathe says.
The researchers plan to use this technique next to study what happens to synapses when they block the expression of genes associated with specific diseases, in hopes of developing new treatments that could reverse those effects.
Do you have a unique perspective on your research related to neuroscience? The Science Advisory Board wants to highlight your research. Contact the editor today to learn more.
Copyright © 2019 scienceboard.net






