March 24, 2022 -- Single-cell sequencing techniques offer an unprecedented level of resolution in distinguishing the many different cell types that make up our organs. However, this comes at the cost of losing information on how tissues are structured. Now, in new research published on March 21 in the journal Nature Biotechnology, researchers have described an approach to combine single-cell sequencing experiments with spatial transcriptomics to create maps of tissues according to cell type.
Our bodies contain hundreds of different types of cells, each specialized for its role. Some, like neurons, are instantly recognizable under a microscope, while others that may appear identical at first glance actually function very differently at a molecular level. Sequencing the RNA transcripts produced by these cells provides scientists with a glimpse into which genes are active and what the cells are doing.
Over the last decade, RNA sequencing has become so sensitive that it is possible to study individual cells. Compared to analyzing bulk samples containing a mixture of cell types, a single cell approach allows researchers to identify activity driven by only a subset of cells. This has proven invaluable for discovering rare cells in tissues and revealing the variety of cell types and states in tumors. However, for this technique to work, the tissue must be broken down into individual cells, meaning that all knowledge about how the cells are structured in the tissue is lost.
Spatial sequencing methods, by contrast, provide information on the tissue structure but do not identify individual cell types. This approach works by sequencing the RNA found in slices of tissue that contain a mixture of cells. Comparing the results across sections makes it possible to detect changes through different parts of the tissue structure. Techniques to "deconvolute" the spatial data can infer the proportion of cell types in each slice but cannot provide single-cell resolution.
Now, researchers have created a new computational approach called CellTrek that combines the data from the two methods to create a map of tissues according to individual cell types.
"Single-cell RNA sequencing provides tremendous information about the cells within a tissue, but, ultimately, you want to know where these cells are distributed, particularly in tumor samples," said senior author of the paper Nicholas Navin, PhD, professor of genetics at the University of Texas MD Anderson Cancer Center. "This tool allows us to answer that question with an unbiased approach that improves upon currently available spatial mapping techniques."
Understanding tumor structure
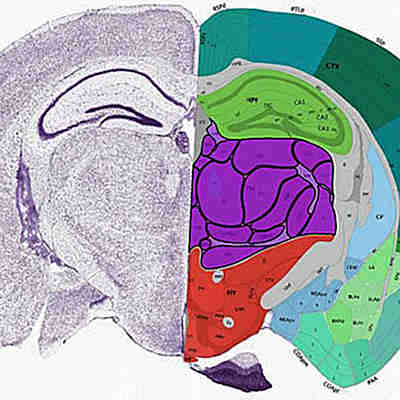
The team used CellTrek on existing datasets of mouse brain and kidney tissues, showing that the technique could detect patterns in the structure of different cell types and states.
They then gathered new data from two samples of ductal carcinoma in situ (DCIS), which is considered the earliest form of breast cancer. These cancerous cells have a low risk of spreading to other regions, but it is difficult to predict in which cases this will happen, so many women undergo treatment as a precaution. A better understanding of how DCIS becomes invasive could help to spare treatment in cases where it may never cause harm.
The researchers analyzed 6,800 single cells and 1,500 spatial transcriptomics spots in one DCIS sample. CellTrek identified three subgroups of cancer cells that were separated in different regions of the ducts. These had different patterns of active genes, which could influence how they respond to treatment.
In the other sample, CellTrek was used to study the arrangement of immune cells surrounding the tumor. This identified several types of immune cells and found that those closest to the cancer had suppressed activity.
Allowing deeper classifications
"While this approach is not restricted to analyzing tumor tissues, there are obvious applications for better understanding cancer," said Navin. "Pathology really drives cancer diagnoses and, with this tool, we're able to map molecular data on top of pathological data to allow even deeper classifications of tumors and to better guide treatment approaches."
The team also developed additional modules to analyze the CellTrek results further. These allowed the researchers to show which cell types are more likely to be found together in the tissue, which could help to study how cells communicate with each other, and how gene expression patterns change across tissue slices.
CellTrek could be improved further, the authors suggest, by including image recognition or deep-learning approaches to identify cells. The computational technique could also be adapted to include other types of data, including epigenetic regulation, which would help research in developmental biology and cancer.
Copyright © 2022 scienceboard.net