November 6, 2019 -- A research team at the University of California, Riverside, has discovered the structure of a novel RNA-modifying enzyme and identified the mechanism that controls substrate specificity. The study was published in Nature Communications on November 6 and explains how protein machinery in cells is regulated to target RNA molecules for modification.
N6-methyladenosine, otherwise known as m6A, is the most abundant modification in mRNA. Zinc Finger CCHC-Type Containing 4 (ZCCHC4) is a RNA m6A methyltransferase (MTase) that methylates human 28S rRNA. This zinc finger is highly conserved in multicellular organisms. Moreover, ZCCHC4 influences cell proliferation and has been linked to cancers. Particularly, it is overexpressed in tumors associated with hepatocellular carcinoma -- the most common type of primary liver cancer.
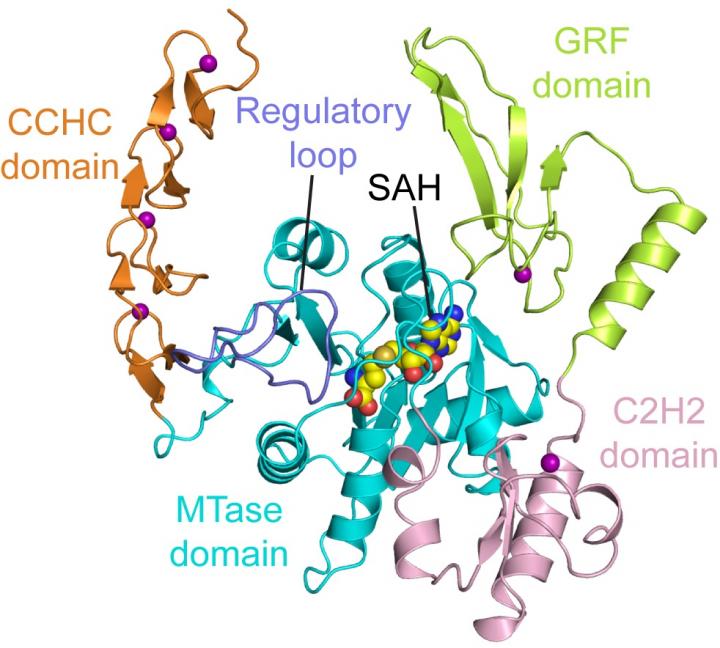
The m6A modification has received enormous attention in recent years due to the important role it plays in RNA metabolism and biology. How it is dynamically programmed and distributed in cells, however, remains poorly understood. The team suggests that ZCCHC4 could be a crucial component of this modification process.
The 3.1 å-resolution crystal structure of ZCCHC4 was determined by crystallization. For successful crystallization, the researchers produced an enzymatically active and structurally rigid fragment by adding alanine, then complexed the structure with S-adenosyl-L-homocysteine (SAH). Subsequently, the crystals were purified and diffracted using X-rays then analyzed with iterative models to determine the structure.
"This is the first time anyone has determined the crystal structure of ZCCHC4," said Jikui Song, an associate professor of biochemistry at UC Riverside. "Our discovery can be used for structure-based drug design against cancers and lead to a better understanding of how m6A, a modification associated with numerous biological processes, is installed on ribosomal RNA."
In the current study the researchers found that ZCCHC4 forms a multidomain RNA-binding platform, permitting its specific recognition of a stem-loop feature inherent in 28S rRNA. The structure of ZCCHC contains four closely packed domains: an N-terminal GRF zinc finger domain is followed by a C2H2 zinc finger domain, an MTase domain, and a C-terminal CCHC zinc finger domain, folded into an integrated structural unit. Furthermore, analysis revealed a loop segment that controls both the enzymatic catalysis of ZCCHC4 and its SAM-binding activity, providing a potential mechanism by which ZCCHC4 achieves substrate specificity.
"The structure of ZCCHC4 provides an understanding of how this enzyme is wired to specifically act on 28S ribosomal RNA,'" Song said, noting a ribosome is assembled with differently sized subunits. "We now understand that this enzyme is controlled by an 'autoinhibitory' mechanism that has been observed in many other cellular processes."
Next, the research team will continue to explore how various DNA and RNA modifications in cells are created, which has strong implications in health and diseases.
Do you have a unique perspective on your research related to biochemistry or moleculear biology as it pertains to drug discovery? Contact the editor today to learn more.
Copyright © 2019 scienceboard.net



