February 15, 2022 -- Researchers have used spatial transcriptomics (ST) to uncover an unprecedented view of the molecular regionalization of the murine colon. This research, published in Nature Communications on February 11, provides novel insights into inflammatory bowel disease (IBD).
The intestine relies on the constant regeneration of the intestinal epithelium to maintain homeostasis. Breakdown in regenerative pathways may lead to the development of pathologies, such as IBD. Therefore, the intestinal barrier must quickly adapt to promote tissue regeneration and healing following injury.
However, the cellular and molecular circuitry at steady-state conditions and how it adapts to challenges is yet to be fully characterized. When an intestinal barrier injury occurs, damaged epithelial cells are shed and are replaced by mobilizing intestinal stem cell (ISC)-derived cells. This phenomenon, however, is highly dependent on the microenvironment.
ST is an unbiased technology allowing sequencing of polyadenylated transcripts from a tissue section. In the current study, a team from Karolinska Institute and University Hospital in Sweden, in collaboration with other research groups, systematically compared ST of colonic tissue under steady state and upon mucosal healing to identify and spatially map transcriptional signatures of tissue-repair processes, immune cell activation/recruitment, proregenerative pathways, and tissue remodeling.
The authors used ST to characterize colonic molecular compartmentalization in the steady-state colon and lymphoid clusters within the tissue. They treated wild-type mice with dextran sulfate sodium in drinking water for seven days, followed by seven days of recovery. At day 14, colonic tissue was taken to generate ST.
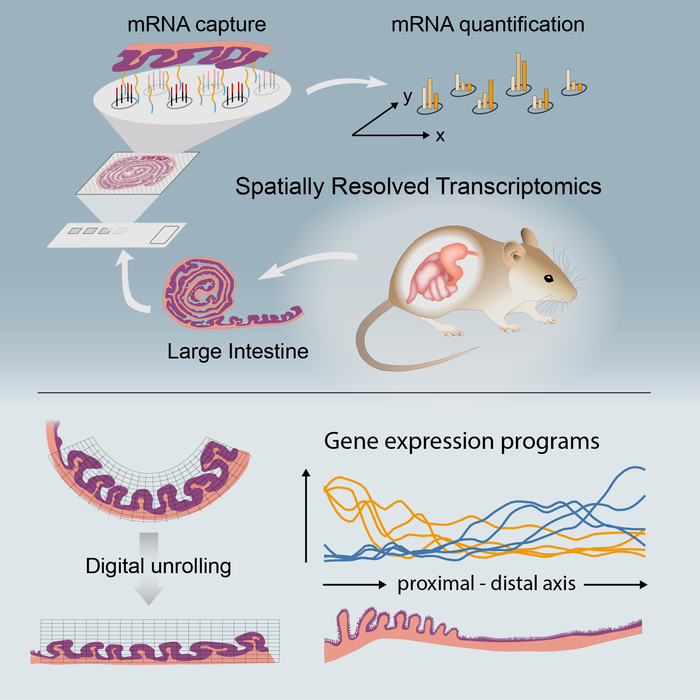
Comparing the steady-state colon with a recovering colon highlighted the differential regulation of cellular pathways and unveiled a negative correlation between p53 (tumor suppressor protein) activity and proliferating epithelial stem cells. Integration of human datasets with mouse ST showed ST within the colonic tissue appeared to be conserved between species, and it highlighted murine ST as a valuable platform for exploring and translating findings on distribution patterns of cells/genes within a tissue.
Ulcerative colitis (UC) 1 and 2 are the two subgroups of IBD patients, characterized by comparable histological disease severity but with different underlying genetic signatures. The study's authors demonstrated that UC1 patients may possess higher tissue damage and ulceration compared with UC2 patients, which might contribute to their poor response to biological therapies. Further, they identified IBD risk genes, which will be key in developing therapeutic strategies for IBD patients.
"The gene map can be used to see where in the colon human cells are active, which can make a significant contribution to the development of new treatments and drugs," said corresponding author, Eduardo Villablanca, PhD, the Wallenberg Academy Fellow in Medicine and an associate professor at the Karolinska Institute, in a statement. "We now want to use the same method to create a similar atlas for all digestive organs, from the mouth to the rectum. Our aim is to create a reference map for the gene expression of all these tissues."
Copyright © 2022 scienceboard.net


