August 18, 2021 -- Scientists have developed a new laboratory protocol and bioinformatic pipeline using a long-read sequencing platform from Oxford Nanopore Technologies (ONT) for exploring the composition of circular RNA (circRNA). The technique, published on August 10 in Nature Communications, showed that a group of microexons -- exons smaller than 30 nucleotides -- that had been previously linked to autism are preferentially found in circRNA.
CircRNA is a type of single-stranded RNA which, unlike linear RNA, forms a covalently closed continuous loop. Moreover, circRNA is noncoding but still plays a functional role in a wide variety of cellular processes.
Only a small portion of RNA is circular RNA (about 1%), and scientists still have a limited understanding of circRNA biology, including its biogenesis, regulatory expression, and functions in mammalian cells. In the central nervous system, circRNAs seem to be involved in brain development and inflammation, and they are known to function as biomarkers in cancer. One particular circRNA, ciRS-7/CDR1-AS, has been shown to cause cognitive changes when removed from the brain of a mouse.
One of the challenges for understanding circRNA composition is that until now, it had only been possible to describe circRNA using sequencing techniques that read short stretches of RNA, a limitation imposed by the current state of technology.
The new approach, in contrast, offers a way to conduct long-read sequencing of circDNA. The researchers applied the Oxford Nanopore platform's long-read sequencing technology to describe the exon (the coding portion of a gene) composition of full-length circRNAs in human and mouse brain tissue samples, as well as the corresponding mRNA.
To zero in on circRNA, the group, led by Karim Rahimi, PhD, of the department of molecular biology and genetics at Aarhus University in Demark, conducted an enrichment protocol in which total RNA from mouse or human brain tissue samples was treated with RiboZero to remove ribosomal RNA and with RNase R to remove linear RNA. An additional polyadenylation step, followed by poly(A) depletion, was performed to ensure removal of all linear RNA.
Next, the researchers applied "gentle" hydrolysis to the circRNA-enriched pool prior to long-read sequencing (circNick-LRS), followed by 3′ end polyadenylation to align with the standard ONT protocol.
After performing long-read sequencing with circNick-LRS, the researchers were able to characterize 18,266 circRNAs in the human brain and 39,623 in the mouse brain. In contrast, short-read sequencing detected 3,740 human circRNAs and 5,299 mouse circRNAs.
Of the circRNAs detected by circNick-LRS, 5,537 were found in both human and mouse brain samples, amounting to 30.3% and 14% of the detected human and mouse circRNAs, respectively. This is on a par with previous studies and shows that the method is reliable in detecting circRNA, according to the authors.
The sequencing results showed that circRNAs, unlike linear RNAs, exhibit a large number of splicing events, such as novel exons, intron retention, and microexons. The authors speculated that the microexons in circRNAs could function as transcription regulators or binding sites for proteins or for microRNA.
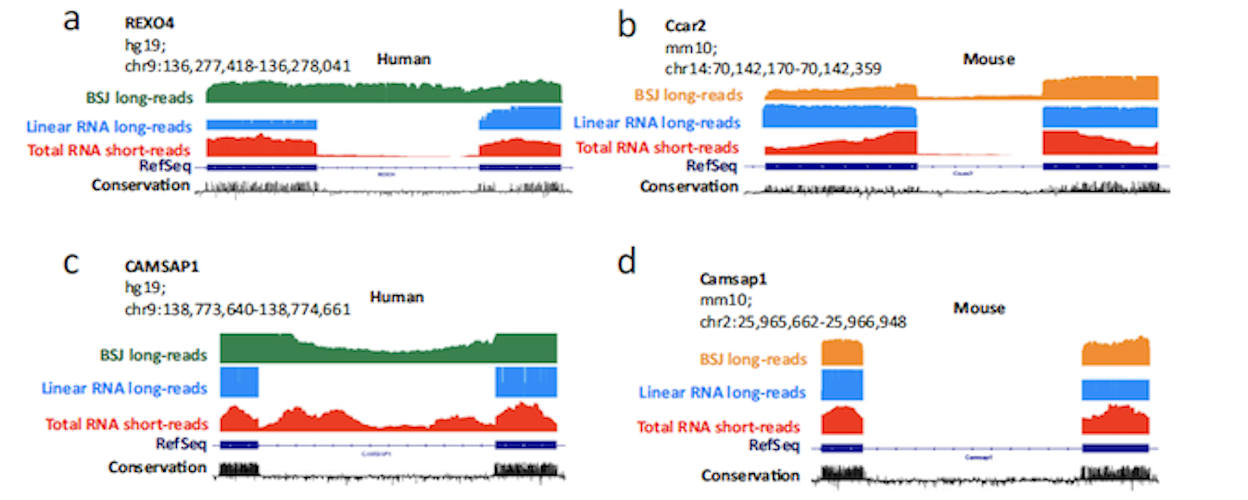
Intriguingly, previous research found that many of the circRNA-associated microexons identified in this study are implicated in brain development, behavior, and autism spectrum disorder, raising the possibility that circRNA-specific RNA elements could play a role in neuronal disease.
The authors noted that in previous studies, these autism-linked circRNA microexons had been misattributed to linear mRNA transcripts, an error resulting from the reliance on short-read RNA sequencing data for the microexon annotation. However, the long-read sequencing results demonstrated that the microexons were associated with circRNA generated from the host gene.
In addition to circNick-LRS, the researchers developed a related technique, circPanel-LRS, that selectively targets a selected set (panel) of circRNAs for long-read sequencing. Using circPanel-LRS makes it possible to get a more detailed view of the splicing landscape of specific circRNAs of interest.
Do you have a unique perspective on your research related to genomics? Contact the editor today to learn more.
Copyright © 2021 scienceboard.net


