May 6, 2021 -- A new study reveals how tiny forces between T cells and their targets can help to jumpstart protective immune responses. Research published in Nature Communications on May 4 uses cutting edge microscopy techniques to track the association and dissociation between T cells and antigens.
T cells are crucial to adaptive immunity because of their ability to detect unique antigens (displayed peptide fragments on the surface of antigen-presenting cells). During this process, T cells and antigen-presenting cells come into contact, through binding of T-cell antigen receptors (TCRs) and peptide/major histocompatibility complex (pMHC) complexes (antigens).
TCRs can detect and bind with moderate affinity to a single antigen among millions of structurally similar molecules. Despite the importance of these interactions, the molecular basis for the recognition process is not well understood.
"Each T cell recognizes one specific antigen particularly well," explained Johannes Huppa, PhD, biochemist and immunology professor at Medical University of Vienna, in a statement. "To do so, it features around 100,000 TCRs of the same kind on its surface."
During infection, infected cells produce fragments of viral proteins on their surface. T cells in the body search for these cells and scan for the presence of antigens that precisely match the TCR. When this match happens, the TCR and antigen can form a somewhat stable bond.
"This works according to the lock-and-key principle," Huppa noted. "For each antigen, the body must produce T cells with matching TCRs. Put simply, each T-cell recognizes only one specific antigen to then subsequently trigger an immune response."
How stable is the binding between antigen and receptor?
In a joint project funded by the Vienna Science and Technology Fund and the Austrian Science Fund, researchers investigated the mechanical processes that take place when an antigen is recognized.
Using a single-molecule Förster resonance energy transfer (FRET)-based live-cell-imaging assay, the team determined that as T cells move away from TCRs within the immunological synapse, they apply only tiny amounts of force -- within 4-9 pN (around 5 x 10-12 N). This is sufficient to break the bonds between the TCR and antigen and helps scanning T cells interact with appropriate antigens.
Testing the strength of the bond
"Let's say we wish to find out whether a surface is sticky - we then test how stable the bond is between the surface and our finger," said Gerhard Schütz, PhD, professor of biophysics at Vienna University of Technology. "We touch the surface and pull the finger away until it comes off. That's a good strategy because this pull-away behavior quickly and easily provides us information about the attractive force between the finger and the surface."
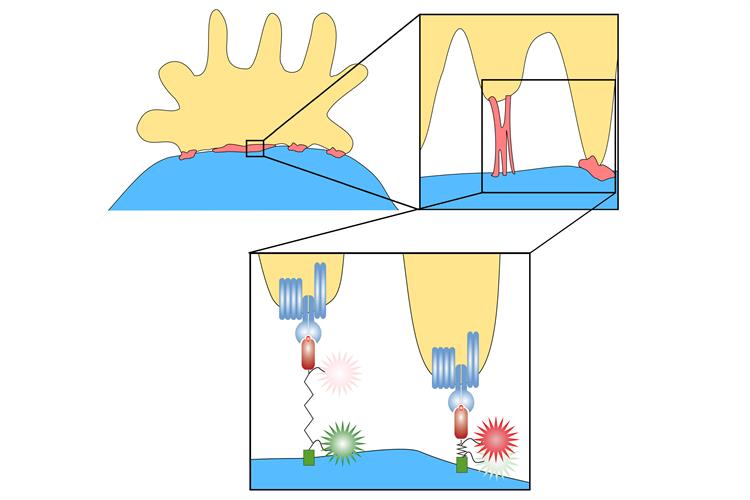
T cells work under a similar principle. Scanning T cells are constantly in motion, and their cell membranes -- which contain TCRs – continuously exert low forces as they test antigen-presenting cells to see if they are a match. During probing, if the T cell does not become activated (i.e., no match), the TCR-antigen bonds require less force and quickly dissociate.
When a TCR binds to a matching antigen, a series of molecular events causes formation of TCR microclusters to help initiate immune responses. In this case, after delayed onset (interaction time of a number of seconds), the cell exerts a steadily increasing pulling force until the binding eventually breaks. Pulling force measurements can provide information about whether it is the antigen that the cell is looking for.
"This process can actually be measured, even at the level of individual molecules," said Janett Göhring, PhD, an author of the study and researcher at both Medical University of Vienna and Vienna University of Technology.
The researchers used total internal reflection fluorescent structured illumination microscopy (TIRF-SIM) to examine the organization of TCR microclusters providing high resolution in the spatial and temporal context.
"The more traction is exerted on the protein, the longer it becomes, said Lukas Schrangl, PhD, first author and researcher at Vienna University of Technology. "With special fluorescent marker molecules, you can measure how much the length of the protein has changed, and that provides information about the forces that occur."
They determined that there are two spatially distinct domains composed of zeta-chain-associated protein kinase 70 (ZAP70)-bound TCR and linker for activation of T cells (LAT)-associated signaling complex. TCR-antigen kinetics revealed delayed recruitment of ZAP70 and associated signaling molecules and distinct patterns of TCR dissociation during T-cell activation.
"Understanding the behavior of T cells at the molecular level would be a huge leap forward for medicine," Huppa said. "We are still leagues away from that goal."
But the authors noted that the current study goes a long way in showing that chemical and mechanical effects play a role in T-cell activation and should be considered together.
Do you have a unique perspective on your research related to immunology? Contact the editor today to learn more.
Copyright © 2021 scienceboard.net


