March 18, 2021 -- A new study finds that the enhanced infectivity of the dominant G614 variant of the SARS-CoV-2 virus largely results from the increased stability of its spike protein. Findings from the study were published in Science on March 16.
The SARS-CoV-2 spike (S) protein is processed by a furin-like protease into the receptor-binding fragment S1 and the fusion fragment S2. Engagement of the receptor-binding domain (RBD) in S1 with the viral receptor angiotensin converting enzyme 2 (ACE2) on the surface of the host cells is followed by a second proteolytic cleavage within S2.
As a result, the S protein undergoes large conformational changes, resulting in the dissociation of S1 and irreversible refolding of S2 into a postfusion structure. This induces fusion of the virus and host cell membrane to initiate infection.
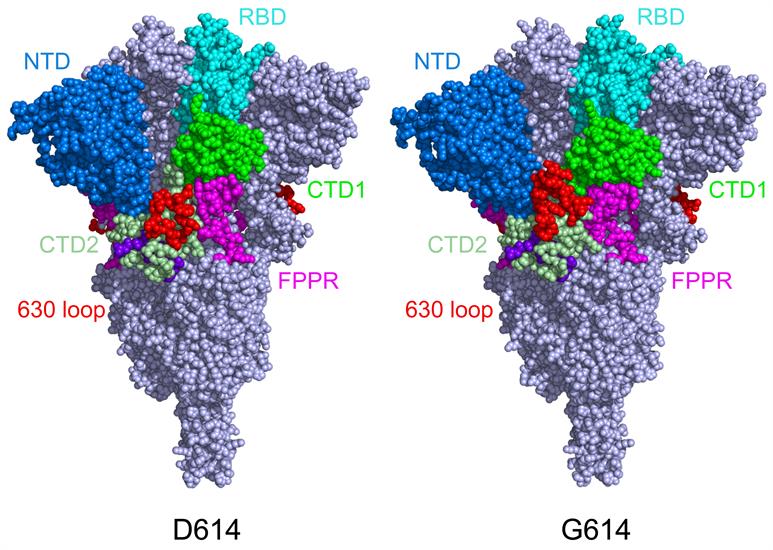
In the prefusion spike protein structure, the S1 subunit folds into four domains -- the N-terminal domain, RBD, and two C-terminal domains -- and wraps around the prefusion S2 subunit. The RBD can take two distinct conformations: "up" for the receptor-accessible state and "down" for the receptor-inaccessible state.
D614G prevents spike shape changes
A variant of SARS-CoV-2 with a single-residue substitution (D614G) in its spike protein has rapidly become the dominant strain throughout the world. Since this mutation occurred, it has further evolved into variants of concern, including the fast-spreading U.K., South Africa, and Brazil variants.
In the new study, researchers from Boston Children's Hospital researchers led by Bing Chen, PhD, analyzed how the structure of the SARS-CoV-2 spike proteins changed with the D614G mutation. The team imaged the spike protein of the D614 "original" spike and the G614 variant spike using cyro-electron microscopy to visualize atomic-level crystal structures.
Chen's team previously reported that D614 spike proteins sometimes change conformation and dissociate before the virus gets a chance to bind to cells. When the team compared eluted proteins of D614 to those of G614, they found that the original spike can be found in all conformations, including prefusion trimers, postfusion trimers, and dissociated monomers, whereas the G614 spike protein was primarily found in the prefusion form.
"Because the original spike protein would dissociate, it was not good enough to induce a strong neutralizing antibody response," Chen said.
The team found that the S1 subunit dissociated more readily from the D614 virus than the G614 virus, suggesting that the D614 viral spike protein is less stable than the G614 variant. Specifically, the G614 trimer was found in the RBD-up conformation (accessible) more frequently than the D614 trimer.
Intriguingly, they also observed that the G614 variant binds more weakly to the ACE2 receptor than the D614 trimer. The researchers explained that the stability of the G614 spike proteins still renders the virus overall more infectious.
"Say the original virus has 100 spikes," Chen explained. "Because of the shape instability, you may have just 50% of them functional. In the G614 variants, you may have 90% that are functional, so even though they don't bind as well, the chances are greater that you will have infection."
The researchers confirmed that the D614G substitution eliminates a salt bridge between D614 in the second C terminal domain of one subunit and K854 in the fusion peptide proximal region (FPPR) of the adjacent subunit, but the inaccessible "closed" conformation remains structured, adding to the stability of the spike protein.
They attributed the stability to the ordered nature of 630 loops (a region that neighbors the C terminal domains). This causes the spike trimer to form more often the one-up RBD conformation (S1 subunit is blocked from dissociating) because the two other RBDs remain inaccessible due to the structure of the 630 loops. This lowers the overall ACE2 affinity of the G614 trimers.
Future directions with the G614 variant: vaccines
In the paper, Chen proposed that redesigned vaccines incorporate the code for this mutant spike protein. The SARS-CoV-2 S protein has been the target of most first-generation vaccines, almost exclusively using the D614 sequence. However, as the G614 mutation has become dominant, these vaccines are constrained and may not target the prefusion form of the spike protein.
The authors suggested that the G614 spike trimer is a superior immunogen for eliciting protective neutralizing antibody responses. They add that it would be an excellent scaffold for designing next-generation vaccines against new variants.
The Boston Children's team is also further applying structural biology to better understand how SARS-CoV-2 binds to the ACE2 receptor, with an eye toward therapeutics to block the virus from gaining entry to host cells. They are exploring potential "decoy" ACE2 proteins that bind the virus more strongly than natural ACE2 found in human cells.
Do you have a unique perspective on your research related to infectious diseases or virology? Contact the editor today to learn more.
Copyright © 2021 scienceboard.net


