April 28, 2020 -- A new taxonomic classification system for viruses has been proposed by the International Committee on Taxonomy of Viruses (ICTV), the group that oversees the official classification of viruses and naming of taxa. A consensus statement on the proposal appears in Nature Microbiology.
Following their discovery in the 19th century, viruses were traditionally grouped by relative relatedness and how they cause disease. However, recent advancements in technology have allowed researchers to gain a deeper appreciation of the importance and distribution of viruses beyond the original parasitic-pathogen model. Moreover, a technology called shotgun metagenomic sequencing has revealed how diverse viruses are in the environment and in healthy organisms.
To understand the true extent of virus genomic diversity and the origins and forces that shape their diversity, virologists needed to systematically rationalize viral relationships that reflect their macroevolution and develop a framework that accommodates the multiorigin nature of viruses.
Five-rank structure -- A step in the right direction
In its earliest efforts, the ICTV classified viruses into taxa of only genera and families. Over time, the classification scheme expanded to include a five-rank hierarchy of species, genus, subfamily, family, and order. This structure matched a section of the Linnaean hierarchical structure used in the taxonomies of cellular organisms and remained in place until 2017.
Moreover, viral taxa classification has changed over time from a traditional phenotype-based characterization process to a multistage process that includes extensive genomic characterization and comparative sequence analyses of conserved genes and proteins.
Extending the taxonomy of viruses is difficult given their nature. Lineages are exceptionally tricky to tease out. Unlike cellular life, viruses acquire their genomic material from many sources. This includes horizontal transfer of genetic elements that allows viruses to swap elements, leaving no clear line of descent.
Further, viral mutation rates are much faster and more prolific than in cell-based organisms, owing to poor mechanisms for genomic proofreading and error correction, as well as select pressure for diversification.
A new structure to classify the virosphere
"With viral metagenomic studies (which involve sequencing genetic material directly recovered from the environment), we are discovering large amounts of viruses that we cannot really put into any particular order," explained Arvind Varsani, PhD, molecular virologist with Arizona State University and member of the ICTV Working Group, in a statement. "We were tasked with trying to come up with a better taxonomic framework."
In 2016, the ICTV initiated a discussion to consider how virus taxonomy hierarchy could more completely mirror the Linnaean taxonomy system (species, genus, family, order, class, phylum, and kingdom) and accommodate a virosphere-wide tree (or trees) from the roots to the tips of the branches.
The ICTV proposed a new extended, formal virus classification hierarchy that would provide 15 ranks, including eight primary ranks (species, genus, family, order, class, phylum, kingdom, and realm) and seven secondary ranks. The new ranks capture the large-scale divergence of viruses within the 15 ranks. For example, the newly created Riboviria taxon (a realm) includes all RNA viruses encoding an RNA-directed RNA polymerase.
The new rank hierarchy and changes to the ICTV International Code of Virus Classification and Nomenclature were approved by the ICTV Executive Committee and ratified by the ICTV in 2019.
To demonstrate how the new system can be applied, the ICTV provides taxonomic classifications for two RNA viruses: the Ebola virus (EBOV) and the severe acute respiratory syndrome coronavirus (SARS-CoV), as well as a double-stranded DNA virus, herpes simplex virus 1 (HIV1). Although EBOV and SARS-CoV are both RNA viruses that infect humans, they only match at the realm level.
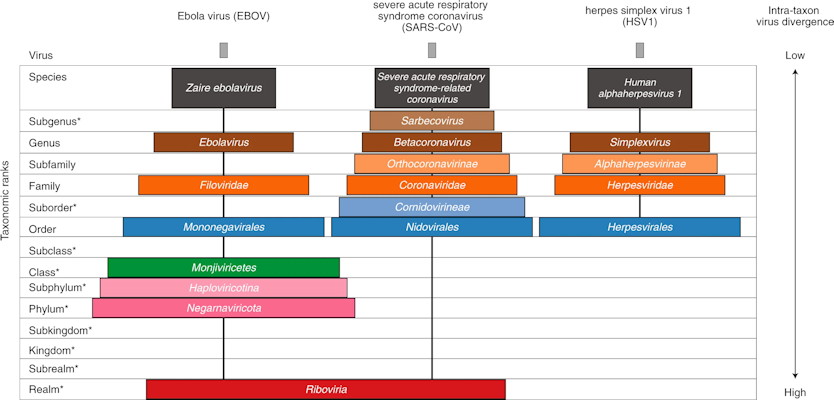
How the new structure can help with SARS-CoV-2
Amidst the chaos of researchers scrambling to understand where and how the novel coronavirus arose, it is essential to properly classify the emergent human pathogen and how it relates to other viruses. This information will inform decisions about surveillance, therapeutic approaches, and public health efforts.
The virus causing the current outbreak of coronavirus disease, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was named after the ICTV determined that the virus belonged to the existing species SARS-CoV, based in part on conserved proteins involved in SARS-CoV-2 viral replication.
Earlier classifications of coronaviruses were largely based on studies of serological activity with viral spike proteins, which give them their characteristic halo-like appearance. But the new taxonomic hierarchy proves that it is capable of situating coronaviruses like SARS-CoV-2 within the enormous web of viruses across the planet, known as the virosphere.
Do you have a unique perspective on your research related to virology? Contact the editor today to learn more.
Copyright © 2020 scienceboard.net






