October 29, 2019 -- Researchers from the University of Bonn propose a new method to determine the prognosis of disease using the patients' tumor tissue. The results were published in Cell Reports on October 29.
Aggressive forms of breast cancer often manipulate the hosts' immune response in their favor. Tumor-associated macrophages (TAMs) are a major component of solid tumor microenvironments. They are involved in cancer-related inflammation and are linked to poor clinical outcomes in several cancers including breast cancer.
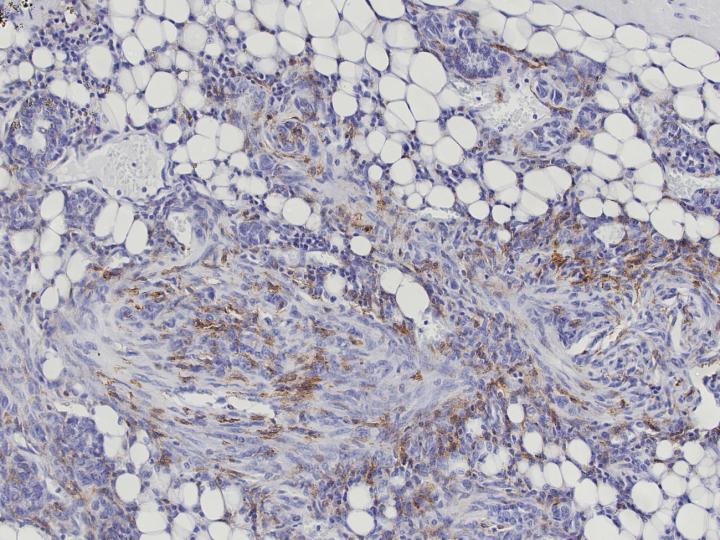
There is a great deal of clinical data available on human cancers, and researchers have used TAM transcriptomes to identify signatures that change in aggressive breast cancer subtypes. But using this information for clinical prognosis has been unsuccessful in the past. Previous approaches have used murine models to define TAM-associated transcriptome signatures, but this is particularly difficult because macrophages possess tissue-specific transcriptional profiles. Therefore, the results of these approaches would reflect tissue-specific differences instead of tumor-specific differences.
The researchers propose an approach to define TAM-specific signatures in murine tumor models by comparison with macrophages from different organs, including the organ of tumor origin. They tested their model with signatures that replicate invasive lobular carcinoma (ILC), the second most common histotype accounting for 10-15% of all breast cancers in humans, and MMTV-NeuT breast cancer model which resembles HER2-positive breast cancers. The gene-signatures from these models can potentially be used as prognostic biomarkers for survival of breast cancer.
The researchers make an important note that the signatures can only be compared to macrophages derived from tissue macrophage counterparts, not by comparison with macrophages derived from unrelated tissues. "Depending on which mutation is responsible for breast cancer, other functions are switched on or off in the macrophages," stresses Thomas Ulas from the LIMES Institute at the University of Bonn.
The researchers validated that murine models were clinically translatable to human forms of cancer. Moreover, they show that the prognostic value of murine models can be evaluated on protein level. "In this case, it was possible to transfer the mouse results directly to humans," explains Joachim Schultze, head of the Genomics and Immunoregulation team at the LIMES Institute. "However, the prerequisite was that the patients suffered from the same form of breast cancer as the animals." The results also demonstrate how important it is to develop specific mouse models depending on the type of cancer.
The data from mice affected by breast cancer was compared with healthy breast tissue. Using state-of-the-art computer algorithms and a bioinformatics approach, the team determined genetic differences between macrophages in the two groups.
In the future this model could lead to the development of new tumor subtype-specific therapies. "However, it will certainly take many years for new treatment options to emerge, if any," says Ulas.
Do you have a unique perspective on your research related to cancer biology? Contact the editor today to learn more.
Copyright © 2019 scienceboard.net






