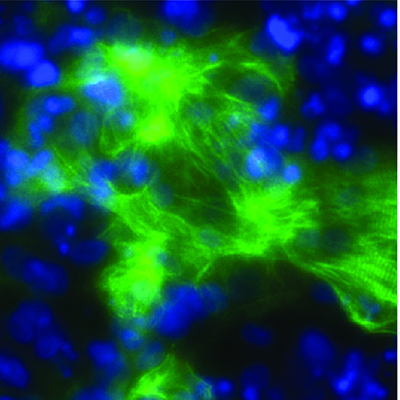
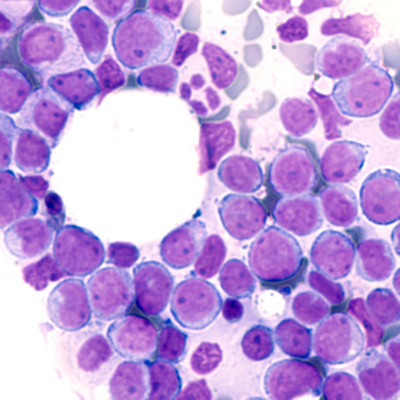
November 3, 2020 -- Researchers have identified a molecule that permits growth of cancers such as leukemia by silencing certain host genes. The results, published in Nature Genetics on November 2, challenge the current understanding of epigenetic control during tumor development.
Post-translational modifications (PTMs) of histones play a significant role in modulating gene expression, which can influence cellular identities during development and cell lineage specification. In cells, the polycomb repressive complex 2 (PRC2) catalyzes formation of H3K27me3, a histone PTM crucial for gene repression and imprinting, epigenomic states, cell-fate determination, and embryogenesis.
H3K27me3-mediated gene regulation is implicated in several disease-associated gene mutations that recurrently target H3K27 or its modifying enzyme, enhancer of zeste homolog 2 (EZH2). These include a missense mutation encoding H3 p.Ly27Met, which specifies a deadly pediatric glioma subtype that displays the altered H3K27me3 pattern. Furthermore, gain- and loss-of-function mutations of EZH2, which encodes the catalytic subunit of PRC2, occur frequently in lymphomas and myeloid neoplasms.
Histone PTMs elicit gene regulatory effects partially through recruiting readers or effectors that control or modulate functional outputs of target chromatin, such as transcription. Established readout models indicated that H3K27me3 is recognized by the chromodomain (CD) harbored within the chromobox (CBX) subunit of polycomb repressive complex 1 (PRC1), which induces chromatin compaction.
However, some genomic targeting occurs via an H3K27me3-independent mechanism that acts upstream of H3K27me3. This raises the question of whether there is another HK27me3 reader other than CBX subunit in mammals.
In a retrospective analysis of published data, a team of researchers from the University of North Carolina (UNC) Lineberger Comprehensive Cancer Center found that the bromo-adjacent homology (BAH) module of the BAHCC1 gene functions as a direct, specific reader of H3K27me3 in mammalian cells.
Structurally, BAHCC1 forms a deep pocket with mixed aromatic and acidic residues that recognize H3K27me3 via a cavity insertion mechanism and recognizes histone tail residues from both the N- and C-terminal regions of H3K27me3. Further, the researchers determined that BAHCC1 is significantly overexpressed in human acute leukemia subtypes and is crucial for malignant growth.
"H3K27me3 is sort of like a gene 'tag' for silencing," said Greg Wang, PhD, senior author and associate professor in the UNC School of Medicine Department of Biochemistry and Biophysics, in a statement. "Our initial interest towards BAHCC1 actually stems from a connection to cancer, in particular, leukemia. And previously, BAHCC1 was little studied. Work on BAHCC1 turns out to be a very interesting journey leading to fundamental understanding of polycomb and gene silencing in general."
Integrated chromatin immunoprecipitation-sequencing and RNA sequencing profiling revealed that BAHCC1 directly binds H3K27me3-demarcated genes in leukemia cells, enforcing gene silencing. Both analyses provide evidence in support of the role of BAHCC1 as a transcriptional repressor. BAHCC1 interacts with transcriptional corepressors, SAP30-binding protein (SAP30BP) and histone deacetylase (HDAC), establishing a molecular basis for BAHCC1-mediated target repression.
The researchers further demonstrated that BAHCC1 plays a role in tumor promotion in acute leukemia using CRISPR-Cas9 loss-of-function techniques. They found that BAHCC1-mediated recognition of H3K27me3 is crucial for optimal gene silencing programs associated with aggressive cancer phenotypes including tumor growth. The function of BAHCC1 relies on an ability harbored within its protein module BAH to scan and directly bind H3K27me3-marked genes.
Targeting excessive H3K27me3-associated signaling is an attractive anticancer treatment strategy and may be achieved through the use of EZH2 catalytic inhibitors, which are under clinical investigation.
According to the authors, the study challenges current theories of CBX proteins as the main effector of H3K27me3 silencing in mammals.
"We now realize that there exists a previously unexplored chapter of important mechanisms that animal cells use for silencing genes," Wang said. "And based on recent research of others, we believe the BAH module and related pathways are evolutionarily ancient and conserved among fungi, plants and animals. In leukemia, BAHCC1 is co-opted to silence tumor suppressive genes and to promote a cancerous program."
Do you have a unique perspective on your research related to cell biology or genetics? Contact the editor today to learn more.
Copyright © 2020 scienceboard.net