June 26, 2020 -- X-ray crystallography performed at room temperature of the main protease of SARS-CoV-2 may be more physiologically relevant than standard low-temperature x-ray structures, according to a June 24 report in Nature Communications.
X-ray measurements help to build 3D structures that can be utilized in supercomputing simulations. Comprehensive 3D models could help find drug inhibitors to block replication of the virus and help end the COVID-19 pandemic.
SARS-CoV-2 employs a chymotrypsin-like protease (main protease) that enables the production of nonstructural proteins that are essential for viral replication. The main protease has been an attractive drug target for specific SARS-CoV-2 protease inhibitors, although none has been approved by the U.S. Food and Drug Administration despite significant research effort during the past 15 years.
Of the three domains in the main SARS-CoV-2 protease, the third domain is helical and plays an essential role in the protease function by forming a functional dimer. The catalytic site of the main protease is buried in an active site cavity on the surface of the protein.
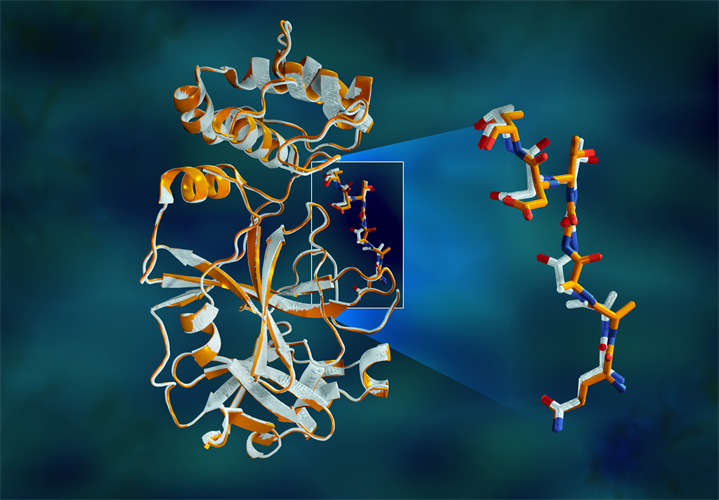
"The protease is indispensable for the virus life-cycle. The protein is shaped like a valentine's heart, but it really is the heart of the virus that allows it to replicate and spread. If you inhibit the protease and stop the heart, the virus cannot produce the proteins that are essential for its replication. That's why the protease is considered such an important drug target," said author Andrey Kovalevsky, PhD, of Oak Ridge National Laboratory (ORNL), in a statement.
Viewing the main protease at room temperature
Many low-temperature x-ray structures of the ligand-free main protease have been used for molecular docking simulations of various small molecules. This methodology is great for detecting heavy elements such as carbon, nitrogen, and oxygen, but samples must be cryogenically frozen (100 K; -280°F) to withstand radiation long enough to collect data.
To minimize radiation damage of the protein, the researchers grew large crystals that could be measured at room temperature using an in-house x-ray machine with a less intense beam than synchrotron x-ray machines. This was the first time that the structure of the main protease had been measured at room temperature, which is closer to the physiological temperatures at which cells normally function, according to the authors.
"Growing protein crystals and collecting data is a tedious and time-consuming process. In the time it typically takes to prepare and ship the sample to a synchrotron, we were able to grow the crystals, take the measurements and begin analyzing the data," said lead author Daniel Kneller, PhD, postdoctoral researcher at ORNL. "And, when there's a pandemic with many scientists mobilizing to study this problem, there's not a day to spare."
The study revealed major disparities between the orientations of the enzymatic backbone and some catalytic site residues in the room-temperature and cryogenic samples. The researchers suggest that freezing the crystals may introduce structural artifacts that could result in a less accurate understanding of the main protease structure.
Therefore, Kneller et al suggest that the conformations observed in room-temperature x-ray crystallography may be more relevant for screening of possible drug candidates for SARS-CoV-2.
Searching for drugs against the SARS-CoV-2 main protease
Now, the results of room-temperature x-ray crystallography of the main protease structure of SARS-CoV-2 are being shared with a research team led by Jeremy Smith, PhD, ORNL director and University of Tennessee governor's chair. His team will conduct molecular drug docking simulations using Summit, the fastest supercomputer in the U.S.
"What researchers are doing on Summit is taking known drug compounds and trying to computationally bind them to the main protease for drug repurposing, as well as looking for new leads into other potential drug candidates," said corresponding author Leighton Coates, PhD, ORNL researcher. "Our room temperature data is being used to build a more accurate model for those simulations and improve drug design activities."
Next, the ORNL researchers will use neutron scattering at ORNL's High Flux Isotope Reactor and the Spallation Neutron Source to further complete the 3D model of the SARS-CoV-2 main protease. This will help them locate hydrogen atoms to better understand the critical role that they play in the enzyme's catalytic function and inform drug design efforts.
Do you have a unique perspective on your research related to biochemistry or infectious diseases? Contact the editor today to learn more.
Copyright © 2020 scienceboard.net