December 9, 2022 -- U.S. Department of Energy and Stanford University researchers have revealed how tiny cellular machines -- called chaperonins -- direct the folding of proteins into building blocks that provide essential cellular scaffolding and transport.
Their study, published on December 8 in the journal Cell, challenges a 70-year-old protein folding theory and has implications for treating protein misfolding-linked diseases, including certain cancers, Parkinson's, Huntington's, and Alzheimer's disease.
Proteins start as spaghettilike strings of amino acids but can't function until they're folded into precise flowerlike shapes. Scientists previously thought the chaperonin TRiC passively provided an environment conducive to folding. However, the protein tubulin can't fold into its final, active shape without TRiC's direct assistance.
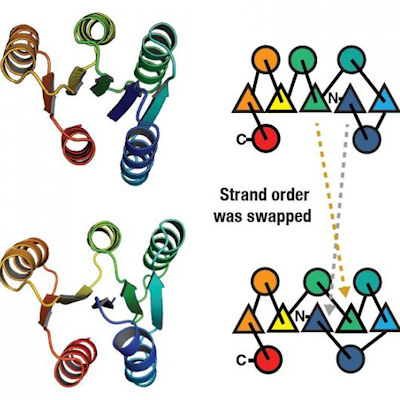
TRiC contains a barrellike nano-chamber that holds proteins inside while they fold. A helper molecule delivers a long tubulin strand into the chamber opening. Then the chamber's lid closes and folding begins. When the lid opens, the finished, folded tubulin leaves. Ten years ago, the team decided to delve into what happens in the TRiC chamber.
Compared to chaperonins' simple folding chambers in bacteria, human TRiC is complicated. Each of the eight subunits presents a distinct surface inside the chamber. As the chamber's lid closes over a protein, electrostatic charges appear on its inner walls. They attract oppositely charged parts of the tubulin strand and "tack" them to the wall to create the proper shape and configuration for the next folding step. Meanwhile, TRiC subunit "tails" dangling from the chamber wall grab the tubulin at specific times and places to anchor and stabilize it.
The researchers captured four distinct steps in the TRiC-directed folding process at near-atomic resolution with cryo-electron microscopy (cryo-EM) and confirmed the sequence with a yearslong series of biochemical and biophysical tests. Eventually, they built a picture of the tubulin's changing shape inside the TRiC chamber during folding, which matched the cryo-EM images.
Seeing proteins folding is "something people have been trying to do for years, and now we have done it," Wah Chiu, PhD, a co-author of the study said in a statement. "This is the most exciting protein structure I have worked on in my 40-year career."
Copyright © 2022 scienceboard.net