August 11, 2020 -- A new study has demonstrated that "foreign" DNA -- DNA transferred horizontally into a species from a source other than a parent -- can become functional over time and can impact an organism's evolution and fitness, according to a paper published August 10 in Nature Ecology and Evolution.
Horizontal gene transfer is the lateral exchange of foreign DNA between unrelated species. Horizontal gene transfer can contribute to the spread of antimicrobial resistance but can also facilitate the engineering of "designer organisms" with desired phenotypes.
Therefore, understanding how these genes from foreign DNA assimilate into a new genome is of interest. The mechanisms of basic gene transfer have been established, but studies on the functionality of gene replacement are insufficient to show how organisms adapt to foreign DNA that initially provides no benefit to the fitness of the organism, but that later becomes functional.
Bioengineers from the Systems Biology Research Group at the University of California, San Diego (UCSD) used CRISPR to generate gene-swapped strains of Escherichia coli with donor DNA from species across all domains of life. The genes pgi and tpiA -- enzymes involved in sugar metabolism in E. coli -- were replaced.
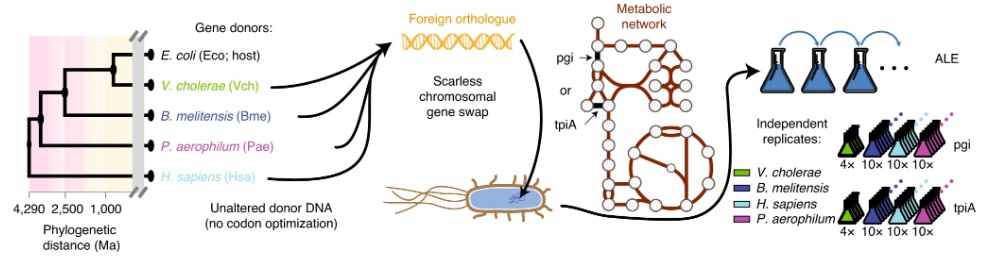
Experimental workflow schematic. Image courtesy of the Palsson Lab.
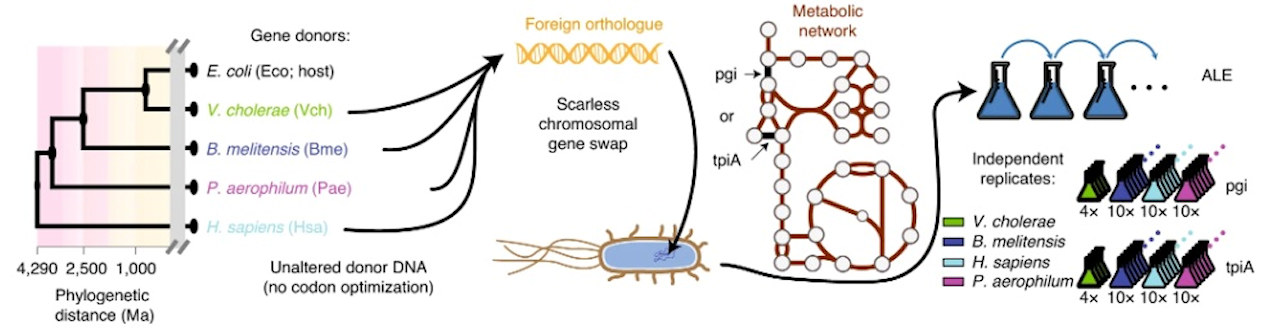
Experimental workflow schematic. Image courtesy of the Palsson Lab.
An automation-enabled large-scale experiment using an "evolution machine" (robotic systems used to study how engineered bacteria adapt to replacement of important genes with foreign versions) generated hundreds of mutant strains that were evolved for more than 50,000 cumulative generations. The experiments were conducted in a matter of months, as opposed to decades if they had been performed manually.
The experiment also allowed cell culture rates to be tracked in real time as populations evolved so that mutant strains could be immediately isolated. With this high temporal resolution of strains, the researchers were able to detect differences in only a single mutation across entire genomes.
Novel ways foreign DNA influences evolution
The researchers found that while E. coli couldn't use the inserted foreign genes immediately, they rapidly evolved and grew just as quickly as before the insertion. For every strain that successfully evolved a use for the foreign DNA, the critical factor was one or more mutations that increased gene expression levels.
Most of the mutations did not occur within the foreign gene but rather in regions that control regulation of the gene -- promoter and ribosome-binding site mutations, especially for pgi swaps. Some of the mutations were shockingly reproducible, occurring independently more than 20 times, indicating that evolutionary outcomes can be predicted to the single DNA base pair.
Of the few mutations that occurred within the foreign DNA (L179L single-nucleotide polymorphism in the upstream gene yiiQ for tpiA swaps), most were synonymous, meaning that the change in codon does not result in a change of encoded amino acid. These mutations are often assumed to have negligible impacts on cell fitness, but the researchers came to a different conclusion based on the results of the study. They found that the mutations were typically located in the stem loop region of messenger RNA (mRNA) causing destabilization of the secondary structure and resulting in more protein to be expressed per mRNA. The authors suggested that this mechanism may be an underappreciated driver of adaptation.
Lastly, the researchers found that many of the evolved strains contained over 90 distinct mutations in the RNA polymerase complex that encodes mRNA transcripts from the DNA sequence. RNA polymerase mutations are common in laboratory evolution experiments; however, the fact that specific mutations clustered into five entirely distinct metabolic regions depending on how the strain evolved is a new finding, according to the authors. This points to the evolutionarily conserved regulatory strategies for rapidly adapting to metabolic perturbations caused by foreign DNA.
"This result shows the importance of systems biology," said Bernhard Palsson, PhD, principal investigator of the study and bioengineering professor at UCSD, in a statement. "Namely, biological function, in this case, is not so much about the parts of the cell, but how the parts come together to function as a system."
Do you have a unique perspective on your research related to genomics or evolutionary biology? Contact the editor today to learn more.
Copyright © 2020 scienceboard.net








