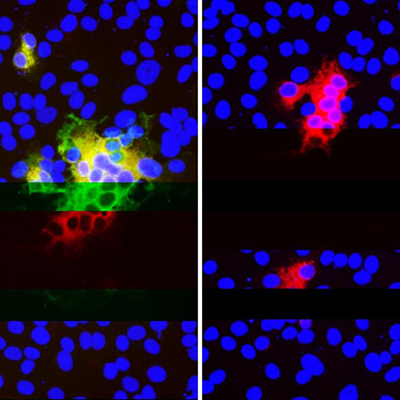
February 16, 2021 -- A new high-throughput proteomics-based strategy to identify tiny antibody fragments -- called nanobodies -- may provide an efficient and effective method for developing therapeutics against the deadly SARS-CoV-2 virus -- including variants. The findings were published in Cell Systems on February 15.
Animals in the camelid family (llamas, alpacas, and camels) produce functional homodimeric and heavy-chain antibodies. Natural antigen-binding fragments derived from these antibodies are called VHH antibodies, or nanobodies.
Antigen engagement is primarily mediated by three complementarity-determining regions (CDR1, CDR2, and CDR3) that form a hypervariable loop to selectively bind targets. In camelids, nanobodies are essential for adaptive immune responses and have recently emerged as promising agents for activation of mammalian humoral immunity.
Nanobodies have several desirable characteristics, including being easily bioengineered. They can be rapidly produced in bulk from microbes, with specific biophysical and structural characteristics. Generally, they are also small in size, stable, and structurally simplistic.
Despite the potential uses of nanobodies, the mechanisms that underlie high-affinity and high-specificity antibody binding have yet to be characterized. This is in part because the industry lacks the appropriate tools and technologies to achieve systematic analysis of antigen-engaged antibody proteomes (the entire set of proteins expressed by the genome under specific conditions).
Using the latest advances in chemistry, biology, and artificial intelligence, University of Pittsburgh School of Medicine (Pitt) scientists developed a new tool that enables the identification and quantification of thousands of high affinity nanobody families.
Furthermore, using advanced proteomics analysis, they were able to map and analyze the epitopes of more than 100,000 antigen-nanobody complexes.
Integrative proteomics to identify high affinity nanobodies
In brief, the researchers immunized camelids with an antigen of interest, followed by the preparation of a complementary DNA library from B cells sequenced by next-generation sequencing.
Concurrently, antigen-specific heavy-chain antibodies were isolated and fractionated. The resulting CDR3 fingerprint peptides were quantified by label-free quantitative proteomics (nanoflow liquid chromatography coupled to high-resolution mass spectrometric analysis). The nanobody fractions were then classified based on binding affinity to the antigen of interest.
Once the researchers fully characterized the structure of the high affinity nanobodies, they worked backward to synthesize the nanobodies as recombinant DNA fragments. High-throughput structural docking, cross-linking mass spectrometry, and mutagenesis were used to verify affinity and to map epitope binding. To explore additional features of high affinity binding, the researchers developed a deep-learning model to identify specific sequences corresponding to high affinity clusters.
"Using this new technique, in a matter of days we're typically able to identify tens of thousands of distinct, highly potent nanobodies from the immunized llama serum and survey them for certain characteristics, such as where they bind to the pathogen," said senior author Yi Shi, PhD, assistant professor of cell biology at Pitt, in a statement. "Prior to this approach, it has been extremely challenging to identify high-affinity nanobodies."
Characterizing high affinity nanobodies
Nanobodies use both shape and charge to enable highly selective antigen binding, which can mimic conserved intracellular protein-protein interactions. Based on the analysis, it appears that nanobodies utilize different CDR combinations to achieve specific, high affinity binding.

The Shi team discovered that the CDR3 region of the nanobodies most significantly contributes to binding affinity. Two main features were identified in high affinity nanobodies. First, significant alterations of charged residues are likely crucial for optimizing the electrostatic complementarity between the paratopes (antigen-binding site) and corresponding epitopes.
Second, high affinity nanobodies often contain an increased number of tyrosines on the CDR3 heads. They also noted that glycine and serine tend to be in the torso (lower specificity region) of CDR3 and provide flexibility to the orientation of the tyrosine side of the binding pocket.
Deep learning revealed a pattern of consecutive lysine and arginine, tyrosine, and glycines as the most informative CDR3 feature for high affinity nanobodies.
SARS-CoV-2 nanobodies: a case study
In the case of most COVID-19 treatments, the spike protein of the SARS-CoV-2 virus is the main therapeutic target. But it has become clear that some sites on the spike are prone to mutations that change its shape and allow for antibody "escape." Shi's integrative proteomics approach can select for binding sites on the spike that are evolutionarily stable and therefore less likely to allow new variants to slip past.
"Most of the vaccines and treatments against SARS-CoV-2 target the spike protein, but if that part of the virus mutates, which we know it is, those vaccines and treatments may be less effective," Shi said. "Our approach is an efficient way to develop therapeutic cocktails consisting of multiple nanobodies that can launch a multipronged attack to neutralize the pathogen."
Utilizing the new tool, the team identified over 8,000 high affinity nanobodies that bind the receptor-binding domain of the SARS-CoV-2 spike glycoprotein. Many of these ultrahigh affinity nanobodies were shown to neutralize SARS-CoV-2 at subnanogram per milliliter concentrations. The nanobodies are easily engineered into multiepitope, multivalent constructs (bacterial cells). This process can produce orders of magnitude more nanobodies compared to the human cell production of traditional antibodies.
"This drastically reduces the cost of producing these therapeutics," said Shi.
In the future, Shi and his team believe the technology could be beneficial for more than just developing therapeutics against COVID-19.
"The possible uses of highly potent and specific nanobodies that can be identified quickly and inexpensively are tremendous," said Shi. "We're exploring their use in treating cancer and neurodegenerative diseases. Our technique could even be used in personalized medicine, developing specific treatments for mutated superbugs for which every other antibiotic has failed."
Do you have a unique perspective on your research related to proteomics or immunology? Contact the editor today to learn more.
Copyright © 2021 scienceboard.net