March 5, 2020 -- Researchers have long known that the uptake systems in clonal cells can differ based on their environments. Scientists reported their observations of this amino acid uptake pathway in bacteria for the first time in a new study published March 5 in Nature Communications.
Auxotrophy is the inability to produce an organic compound essential for growth and is a characteristic common among many bacteria, which often rely on their environment to obtain missing compounds. Auxotrophy shapes interactions between communities of bacterial cells by necessitating syntrophic cross-feeding interactions and the exchange of amino acids. Therefore, it is likely to play a role in determining the composition and stability of microbial communities.
Different kinds of membrane transporters facilitate the uptake of external amino acids, ranging from generic low-affinity transporters that facilitate the uptake of several different amino acids to targeted high-affinity transporters that can import specific amino acids only at high efficiency. Competition between auxotrophic bacteria causes bacteria to sequester amino acids via different systems.
To study this process, microbiologists from the University of Groningen used Lactococcus lactis strains to explore uptake of the amino acid methionine. L. lactis can use two different transporters to import methionine: a high-affinity ABC transporter, called Met-transporter, and a low-affinity transporter, called the branched-chain amino acid permease (BcaP). The low-affinity transporter primarily transports branched-chain amino acids (isoleucine, leucine, valine), but it can also transport methionine and cysteine.
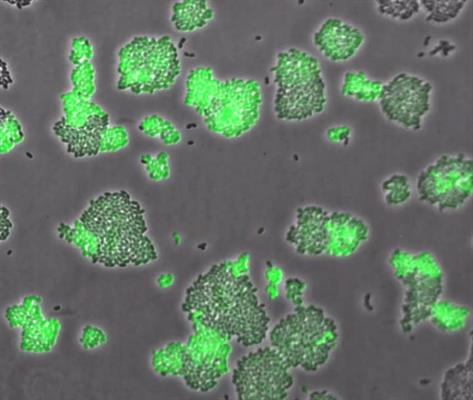
In the study, the researchers reported expression of high-affinity transporter under a range of methionine concentrations. Under methionine-limited conditions, L. lactis differentiates into two phenotypic subpopulations. Moreover, the researchers observed this differential expression stably inherited across tens of generations.
- In the first subpopulation, BcaP is quickly expressed, and upregulation of the low-affinity transporter results in enough methionine to support growth.
- In the second subpopulation, uncharged tRNAMet builds up as a result of methionine starvation, which leads to the upregulation of the Met operon via the T-box riboswitch. This attenuates expression of met RNA and eventually increases expression of the high-affinity transporter to increase the rate of methionine uptake.
The researchers postulate that L. lactis uses long-term phenotypic heterogeneity in a strategy to rapidly adapt to shifted environments without the need for genetic mutation. As the term suggests, it describes different mutations in the same gene that can sometimes give rise to strikingly different phenotypes. In this case, expression of high-affinity Met transporter. According to the authors, this is the first study that shows regulation at the RNA level that can give rise to phenotypic heterogeneity.
Why is methionine uptake heterogeneously expressed?
- Phenotypic heterogeneity could support bet-hedging strategies, in which the population can prepare itself for unexpected changes by expressing both phenotypes. This strategy has been previously reported in L. lactis, where the bacterium prepared to consume alternative future carbon sources.
- Cooperative action occurs between cells with strongly or weakly expressed Met-transporter, in which both cell types benefit. In this metabolic division of labor scenario, metabolites can be exchanged within subpopulations to support the entire community.
"We do know that heterogeneity is a good thing for bacteria," concluded Oscar Kuipers, PhD, a professor of molecular genetics at the University of Groningen, in a statement. "By splitting into two populations, you can better anticipate changes in the environment. That is always a smart strategy."
Do you have a unique perspective on your research related to microbiology or metabolism research? Contact the editor today to learn more.
Copyright © 2020 scienceboard.net






