May 5, 2021 -- Researchers further elucidated how antibodies produced in people who effectively fight off SARS-CoV-2 work to neutralize the part of the virus responsible for causing infection. The study, published in Science on May 4, describes how antibodies targeting sections of the virus outside of the receptor-binding domain (RBD) may be useful in the development of vaccines and therapies.
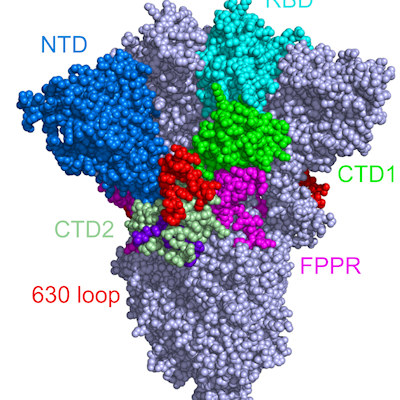
Humoral immunity (antibody-mediated immunity) to the SARS-CoV-2 spike protein, specifically the RBD, can provide protection and as such is the primary antigenic target for most vaccines and monoclonal antibodies. Previous research has focused on antibodies that target the RBD. But antibodies produced by B cells during infection can recognize a variety of spike protein epitopes. The source of the full immunoglobulin G (IgG) plasma antibody repertoire and the epitopes it may target in response to SARS-CoV-2 are not known.
To characterize the IgG repertoire more fully, researchers from the University of Texas at Austin (UT Austin) analyzed blood that was collected during early convalescence from four patients who had been infected with SARS-CoV-2. They determined the lineage composition and relative abundance of constituent IgG antibodies comprising the plasma response to either the intact stabilized spike ectodomain or RBD using the Ig-Seq pipeline, which integrates analytical proteomics of affinity purified IgG fractions with peripheral B cell antibody variable region repertoires.
The researchers found that only four IgG lineages accounted for 93.5% abundance of the total plasma IgG spike ectodomain response. Of these, three lineages bound to non-RBD epitopes (S2 subunit or N-terminal domain). On average, 84% of the plasma IgG targeted epitopes outside the RBD and the most abundant plasma IgG lineage in all donors recognized a non-RBD epitope.
Using enzyme-linked immunosorbent assays (ELISAs) the team confirmed that RBD-binding plasma antibodies included only a minor proportion of all spike-binding IgG in naturally infected individuals.
"We found these antibodies are painting the entire spike, both the arc and the stalk of the spike protein, which looks a bit like an umbrella," said Greg Ippolito, PhD, an author of the study and professor in UT Austin's Department of Molecular Biosciences and the Dell Medical School, in a statement. "The immune system sees the entire spike and tries to neutralize it."
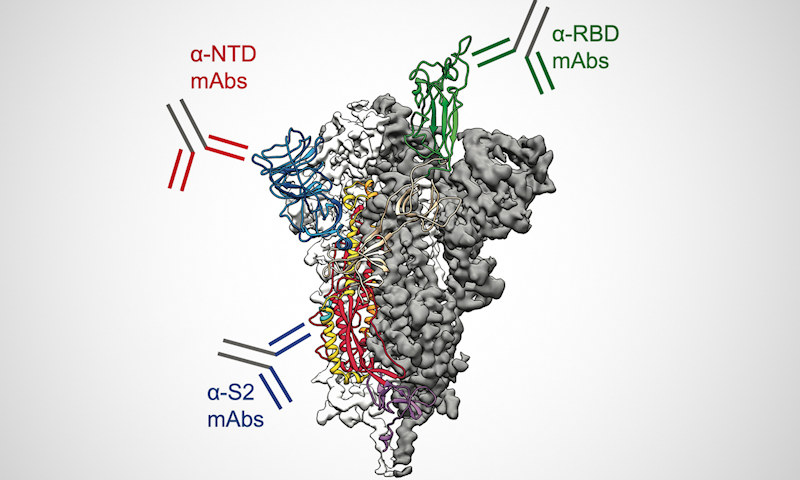
In cell culture, neutralizing antibodies targeting the N-terminal domain (NTD) were able to completely protect against viral challenge. However, the team noted that administration of an antibody cocktail containing several non-RBD plasma monoclonal antibodies was able to provide robust protection and lung viral titers below the limit of detection (LOD) in a high viral load challenge.
Using cryo-electron microscopy, the scientists determined the structure of fragment antigen-binding (Fab) antibody CM25 bound to the trimeric spike ectodomain to study the antibody-NTD interface. Heavy-chain–dominant binding was the main source of interaction, with substantial contacts mediated by interactions between the three complementarity-determining regions (CDRs) and the N3 and N5 loops of the NTD.
The NTD is also a part of the viral spike protein that mutates frequently, especially in several variants of concern. Specifically, mutations in the N3 and N5 loops of the NTD can confer escape from neutralizing anti-NTD antibodies. One reason these variants are so effective at evading the human immune system is that they can mutate around one of the most common and potent types of antibody in the body.
"There's an evolutionary arms race going on between the virus and our immune systems," said Jason Lavinder, PhD, an author of the study and research associate in the McKetta Department of Chemical Engineering at UT Austin. "We're all developing a standard immune response to this virus that includes targeting this one spot and that's exerting selective pressure on the virus. But then the virus is also exerting its evolutionary strength by trying to change around our selective immune pressures."
Despite these maneuvers by SARS-CoV-2, the researchers noted that around 40% of the circulating antibodies target the stalk of the spike protein, called the S2 subunit, which is also a part that the virus does not seem able to change easily.
"That's reassuring," Ippolito said. "That's an advantage our immune system has. It also means our current vaccines are eliciting antibodies targeting that S2 subunit, which are likely providing another layer of protection against the virus."
In terms of vaccines, the authors explained that researchers have a good understanding of what targets they should be including in vaccine boosters or next-generation vaccines against variants of concern.
"It means we have a strong rationale for developing next-generation SARS-CoV-2 vaccines or even a pan-coronavirus vaccine that targets every strain," Ippolito said.
UT Austin researchers are leveraging this information to develop a single coronavirus vaccine to fight infection from all coronaviruses, not just SARS-CoV-2.
Do you have a unique perspective on your research related to virology or infectious diseases? Contact the editor today to learn more.
Copyright © 2021 scienceboard.net