December 1, 2020 -- Transcription factors (proteins that control gene expression) can be used in simple "recipes" to easily convert stem cells into hundreds of different cells and tissues, according to a new study published in Nature Biotechnology on November 30.
Human pluripotent stem cells have nearly unlimited expansion capability and the potential to differentiate into any cell type, however protocols to generate most cell lineages are severely lacking. They often attempt to mimic complex developmental biology timelines and rely on external signals to activate transcription factors to control cell type-specific genetic programs. Ultimately this makes it difficult to generate multiple lineages in the same culture.
An alternative strategy is the direct activation of transcription factors, which can shorten the time for cell conversion. Previous studies applying direct induction of transcription factors for cell fate engineering have used a limited set of developmentally relevant factors or computationally predicted factors from genome datasets with modest accuracy. These "recipes" for conversion are complicated and difficult to implement. With over 1,600 known transcription factors, there is room for improvement in experimental study.
Researchers from the Center for Regenerative Therapies Dresden (CRTD) at Dresden University of Technology (TU Dresden), Harvard University, and the University of Bonn constructed a library of 1,564 human transcription factors to create the Human TFome. The researchers aimed at replacing complicated protocols with simple recipes that use transcription factors without the need for lineage-specific media.
A single transcription factor is sufficient for differentiation
Next, the scientists expressed the Human TFome in human induced pluripotent stem cells (hiPSCs) with a coverage rate of greater than 87% across three hiPSC lines. They screened each transcription factor for its ability to induce differentiation in the three lines using a PiggyBac transposon system. Of the 290 transcription factors that individually induced differentiation, 241 had not been previously associated with cell differentiation.
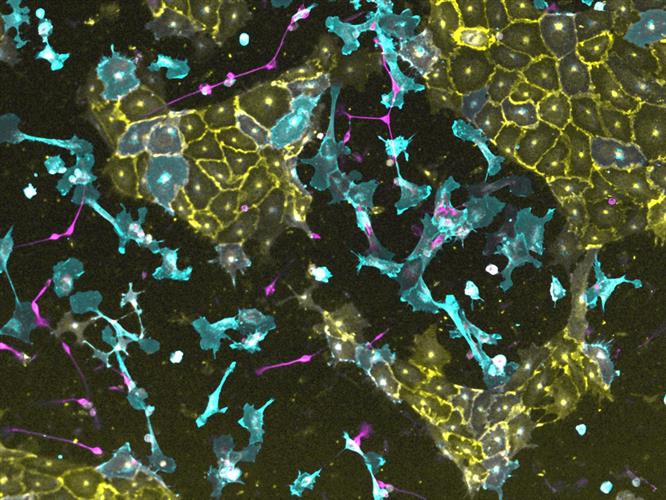
The team then characterized four of the 290 transcription factors in depth - ATOH1, NKX3-1, ETV2, and SOX9. Each of the factors programmed hiPSCs within four days into induced neurons (ATOH1), induced fibroblasts (NKX3-1), vascular endothelial-like cells (ETV2), and induced oligodendrocytes (SOX9). The researchers conducted a variety of in vitro tests to demonstrate that the induced cells were functional.
With goal of constructing complex tissues, the team also investigated the ability of transcription factors to induce parallel programming. They found that the combination of ETV2, NKX31, and ATOH1 in hiPSCs simultaneously generated multiple lineages (neuronal cells, endothelial cells, and fibroblasts) within four days, with approximately equal proportions of each cell type.
Future avenues
The authors envision that the Human TFome would be a powerful tool used in conjunction with other approaches for cell and tissue engineering.
"The advantage of the identified transcription factors is that they are able to convert iPS into body cells particularly quickly and easily and that they can potentially also be used to form more complex tissues," said Dr. Volker Busskamp, PhD, professor at the University of Bonn and the CRTD at TU Dresden, in a statement.
The tool also opens new possibilities in stem cell research.
"The variety, simplicity and speed of stem cell programming using transcription factors makes stem cell research possible on a large scale," explained George Church, PhD, of Harvard. "Worldwide, 50 other groups are already working with our programmable stem cell lines and with the transcription factor library."
Two authors on the paper, Alex Ng, PhD, and Parastoo Khoshaklagh, PhD, of Harvard, have now founded the startup GC Therapeutics, which provides programmable stem cells with customized, integrated transcription factors.
Do you have a unique perspective on your research related to stem cell research or drug discovery & development? Contact the editor today to learn more.
Copyright © 2020 scienceboard.net


