March 3, 2020 -- What's in a name? Possibly a lot, when it comes to determining how to name new virus species based on genetic characteristics. The initial confusion over the naming of the novel coronavirus indicates that the scientific community still has work to do when defining the proper taxonomy of viruses, according to an article published in Nature Microbiology on March 2.
The assessment of virus novelty has implications for virus names and helps to define research priorities in virology and public health. In outbreaks involving newly emerged viruses, appropriate procedures to deal with the viruses need to be established or refined with urgency.
A case in point came with the recent outbreak in humans of a novel coronavirus. The virus was provisionally named 2019-nCoV but later was renamed severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) based on its genetic similarity to the SARS coronavirus identified in 2003.
In the new statement, international bodies are urging researchers to adopt proper referencing to the full taxonomy of coronaviruses. This includes explicit mentioning of the relevant virus species and specific viruses within the species according to proposed naming rules by the International Committee on Taxonomy of Viruses (ICTV).
The naming of RNA viruses needs to consider inherent genetic variability, which often results in two or more viruses with nonidentical but similar genome sequences being regarded as variants of the same virus. Degrees of relatedness address the question of how much difference is large enough to recognize the emerging virus as a member of a new, distinct group. This analysis is addressed in the official classification of virus taxonomy that is overseen by the ICTV.
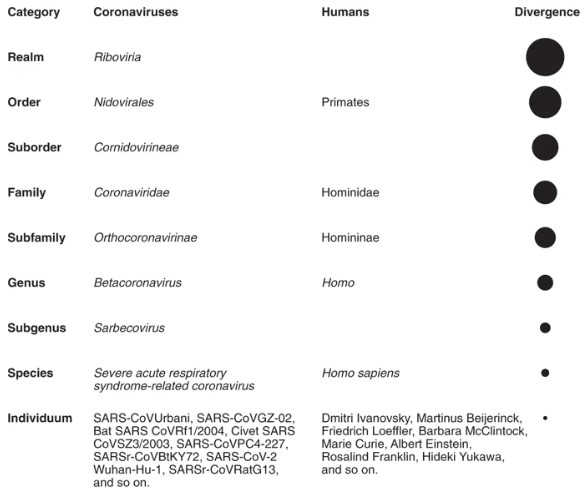
Viruses are clustered in taxa in a hierarchical scheme of ranks in which the species represents the lowest and most populous rank containing the least diverged group of viruses. Study groups within the ICTV are responsible for assigning viruses to higher ranks such as subgenera, genera, and subfamilies.
Specifically, the Coronaviridae Study Group (CSG) is in charge of the classification of viruses and taxon nomenclature of the family Coronaviridae. ICTV also has a responsibility in naming and establishing species. The species name is italicized, starts with a capital letter, and should not be spelled in an abbreviated form (Severe acute respiratory syndrome-related coronavirus). This should not be confused with the convention for virus names, hence severe acute respiratory syndrome coronavirus, or SARS-CoV, as it is widely known.
Defining SARS-CoV-2
The current ICTV classification of coronaviruses recognizes 39 species in 27 subgenera, five genera, and two subfamilies that belong to family Coronaviridae, suborder Cornidovirineae, order Nidovirales, and realm Riboviria.
The CSG uses a computational framework of comparative genomics to quantify and partition the variation in the most conserved open reading frames 1a and 1b of coronavirus genomes. The resulting pair-wise patristic distances (PPDs) demarcate virus clusters at different ranks and taxa.
The group's analysis revealed that SARS-CoV-2 clusters with SARS-CoVs in the species of Severe acute respiratory syndrome-related coronavirus and the genus Betacoronavirus. However, to date, there is no agreement yet on the exact taxonomic position of SARS-CoV-2 within subgenus Sarbecovirus based on varying distance estimates (nucleotide or amino acid measures).
Epidemiological and clinical data for SARS-CoV-2 suggest that the disease and transmission efficiency of the virus is different from SARS-CoV.
The World Health Organization (WHO) is responsible for naming diseases caused by newly emerging human viruses. The WHO recently introduced an unspecific name to denote the disease (coronavirus disease 19 [COVID-19]).
The CSG hopes that by establishing different names for the causative virus (SARS-CoV-2) and the disease (COVID-19), awareness will grow in the general public and public health authorities regarding the difference between the two entities. Uncoupling the naming conventions will support the WHO in efforts to establish appropriate disease names. This is important as next-generation sequencing technologies will provide an increasing number of viruses that do not easily fit the virus-disease model that is widely used.
Therefore, the GSG proposes a naming convention that includes a reference to the host organism that the virus was isolated from, the place of isolation (geographic location), an isolate or strain number, and the time of isolation (year or more detailed) in the format virus/host/location/isolate/date -- for example, SARS-CoV-2/human/Wuhan/X1/2019. They believe that this format will provide critical metadata needed to characterize each particular virus genome. This is required to boost epidemiological and other studies, as well as for public health control measures.
From reactionary to proactive
Traditional public health and basic research efforts have been focused on the detection, containment, and treatment of viruses that are pathogenic to humans after their discovery (a reactionary approach). While the emergence of SARS-CoV-2 into the human population in December 2019 may seem like an independent outbreak from the SARS 2002-2003 event, the two viruses are related and, thus, their evolutionary history and characteristics are mutually informative.
Therefore, the CSG believes that current knowledge of the natural diversity of the species Severe acute respiratory syndrome-related coronavirus is limited and affects the current understanding of the fundamental aspects of the biology of the species and, as a consequence, our abilities to control spread to humans. They suggest an expansion of research that focuses on human pathogens and their adaptation to specific hosts and other viruses in the species.
The group also believes that additional diagnostic tools that target entire species should be developed to complement existing tools to detect individual pathogenic variants (a proactive approach). These steps will provide a reduction of the time required to identify causative agents of novel virus infections in the future and limit the enormous social and economic consequences of large outbreaks.
Do you have a unique perspective on your research related to virology? Contact the editor today to learn more.
Copyright © 2020 scienceboard.net


