July 30, 2018 -- As Next Generation Sequencing (NGS) continues to grow, scientists face many challenges in terms of their data analysis and storage solutions. Our members at The Science Advisory Board have disclosed it is difficult to determine the significance of results as well as store the mass amounts of data. There are many solutions but most lack desired features, creating uncertainty amongst researchers over the best option.
This study highlights the perspective of our NGS using members as we explore critical issues in NGS bioinformatics. Scientists shared Illumina and Thermo Fisher sequencing instruments are by far the most-used NGS platforms, while other market players such as Pacific Bioscience and Oxford Nanopore are gradually entering the space. Interestingly, Roche’s 454 GS series is still used by 11% of our members, though it is now discontinued.

The Science Advisory Board expected that there would be a strong correlation between the leading instrument and awareness of their accompanying informatics packages. From our study, we could conclude Illumina’s BaseSpace solutions, QIAGEN’s CLC Genomics Workbench and Ingenuity Variant/Pathway Analysis solutions, and Thermo Fisher’s Ion Reporter have the highest levels of awareness as well as usage amongst the respondents. We were most surprised to find the majority of commercially available sources of NGS data analysis solutions are “invisible” to our members and they were unaware of certain NGS products.
Our members vary in how they obtain software used for analysis and integration of their NGS data. Regardless, all scientists rely on a mix of internally developed software, freeware distributed throughout the scientific community, commercial packages from small independent vendors, software embedded in analytical instruments and proprietary software designed to run on enterprise solutions. The heterogeneity of the software environment – particularly at large research organization – reflects the rapid evolution of NGS and the difficulty of integrating not only disparate data but also multiple application tools. We found that scientists engaged in basic research are more likely than their academic counterparts to use software developed in-house.
In the past, we have found that scientists’ software decisions are not driven by concerns about installation, documentation or future upgrades and enhancements. The ultimate challenge is the inability to find exactly what they need in a commercial software package. NGS used requirements for software can be unique from user to user, making it difficult to find a universal solution commercially. We believe members would consider a commercial software package if it had customization solutions and post-sales support.
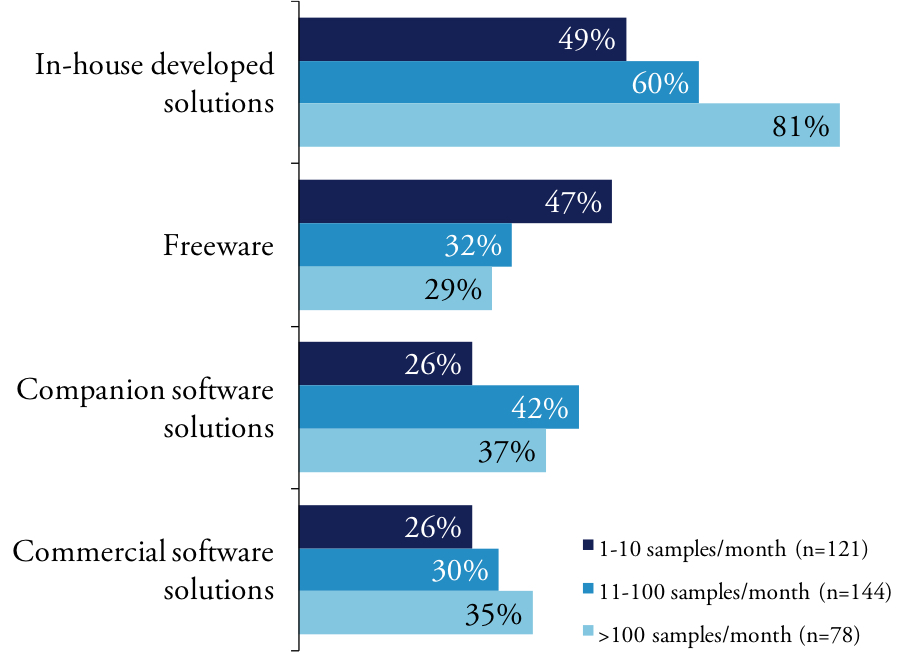
Many researchers using NGS are generating new data in their experiments and not working in silico using data previously collected. It is likely the many scientists are generating the data within the context of their experiment. Our lower throughput users report experiencing more problems and bottlenecks than high throughput labs.
Success in the NGS market will be derived from the ability to foresee the trajectory of future improvements that will result from convergence and the scientific and economic consequences of those improvements. Do you use NGS in your work? Share your thoughts on this topic below!
Copyright © 2018 scienceboard.net


