October 1, 2019 -- A new finding reported in Nature Structural & Molecular Biology on October 1 shows how bacterial transmit motion from an inner motor to an outer tail through a flexible joint in the flagellum. This hook helps researchers understand how bacteria move and allows them to improve therapies against bacterial infections.
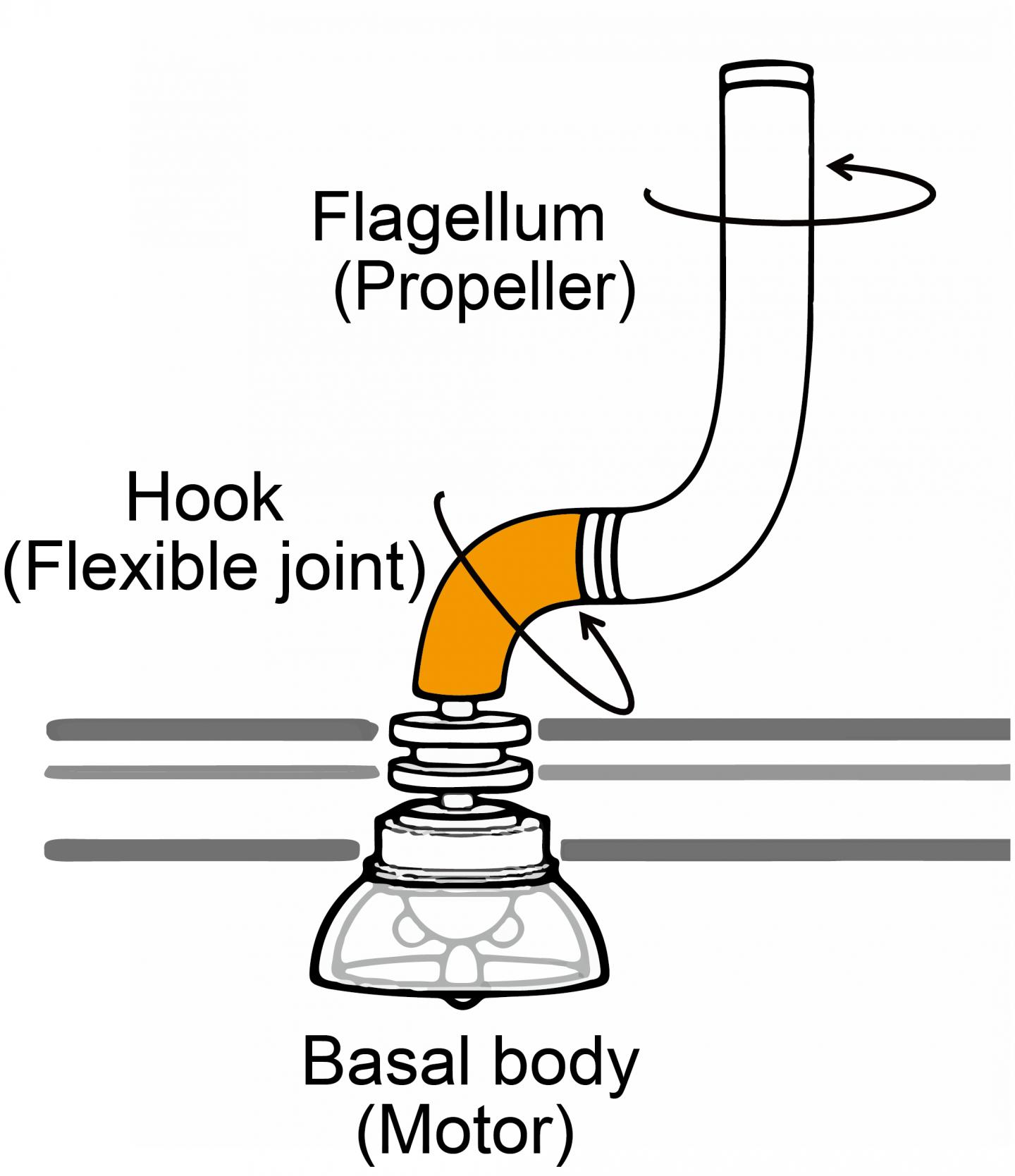
Researchers at Okinawa Institute of Science and Technology Graduate University (OIST) describe how this hook acts as a dynamic joint, transmitting torque enabling bacteria to move. The team used cryogenic electron microscopy (cryo-EM) to show the rigid and flexible co-occurring nature of the hook, despite its single type of protein composition. The hook achieves this through dynamic shifts of subunits, which can adopt at least eleven unique conformations shared along 11 protofilaments.
"While the outer filament is thought to exist in two states, the hook appears to have no set configuration," says Professor Matthias Wolf, senior author on the study. "This contradicts the assumed 'two-state model' thought to explain the hook's function and explains how such a dynamic structure is possible. This intricate system is a testament to evolution's ability to optimize over millions of years."
"The flagellum is an astonishing feat. Its rotor, stator, drive shaft, bushing, bearings, and propeller-like tail synchronize to plunge bacteria forward through viscous fluids at several body lengths per second," says Satoshi Shibata, a lead author on the paper. "In many ways, the hook's helical shape beats human engineering's attempts at creating flexible joints."
In this study, the hook structure was first purified and cooled to cryogenic temperatures for imaging. Cryo-EM was used to generate static images of the hook. Then, three-dimensional maps were reconstructed using OSIT's high-performance computing cluster. The resultant images yield richer insights at near-atomic resolution which allows researchers to consider the relationship between structure and function.
The researchers found that the flagellar hook is composed of approximately 130 subunits, each comprising only a single type of protein. Each subunit is made of three domains which act as rigid bodies. These domains are connected by two flexible hinges. Through imaging, the researchers discovered that the subunits can adopt different conformations, leading them to suggest a polyhook model which can shift during bending.
Moreover, the shifts in conformation allow bacteria to travel from the spinning motor through its helix, eventually rotating the outer filament. The model organism in this study was Salmonella enterica, a human pathogen and major cause of death in the world. Understanding how the flagellum works, and how essential it is for bacterial motility, is the first step in exploiting its function to fight infection, according to the researchers. The team confirmed that this particular motif is conserved in several common bacterial species, suggesting that this is a common feature and ideal therapeutic target.
Do you have a unique perspective on your research related to molecular biology? The Science Advisory Board wants to highlight your research. Contact the editor today to learn more.
Copyright © 2019 scienceboard.net


