April 29, 2021 -- Computer modeling has demonstrated that one of the three mutations of the South African SARS-CoV-2 variant reduces its ability to bind to human cells. The results, published recently in the Journal of Medicinal Chemistry, also suggest that the mutation may help it escape some therapeutic antibodies.
Since its emergence in 2019, several variants of SARS-CoV-2 have raised concerns due to their high transmissibility rates and ability to spread rapidly. Among these new variants are B.1.1.7 (or 501Y.V1) from the U.K., B.1.351 (or 501Y.V2) from South Africa, and P.1 from Brazil.
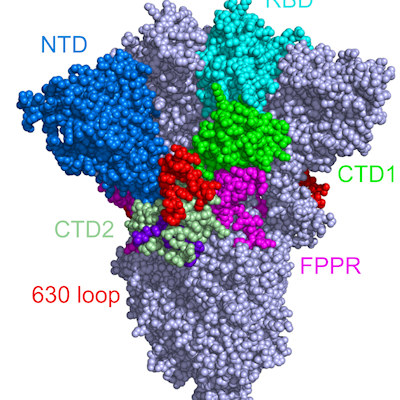
Public health experts and clinicians worry that the variants could undermine current vaccines, antibody therapies, or natural immunity. Indeed, all the variants share some of the same mutations in the receptor-binding domain (RBD) of the spike protein, suggesting that they provide the virus with an evolutionary advantage.
The South African variant bears two mutations (N501Y and E484K) that enhance RBD and human angiotensin converting-enzyme 2 (ACE2) binding. The N501Y mutation does not affect the virus's ability to bind to monoclonal antibodies, whereas the E484K mutation can help the virus escape the therapeutically relevant monoclonal antibodies. However, the significance of the third mutation (K417N; a lysine to asparagine mutation at position 417) is still elusive to the research community.
In a new study, Binquan Luan, PhD, and Tien Huynh from IBM Research wanted to investigate potential benefits of the K417N mutation to the novel coronavirus. They conducted computational analysis of the K417N mutation in the South African variant using molecular dynamics simulations with the goal of gaining a better understanding of its molecular mechanism. The researchers also investigated the effect of the K417N mutation on the RBD's interactions with therapeutic monoclonal antibodies, such as CB6, which recognizes an epitope site in the RBD overlapping the binding site of ACE2.
In the SARS-CoV-2 wildtype RBD, K417 forms a salt bridge with the D30 residue in ACE2 and is important in RBD-ACE2 recognition. The researchers demonstrated that K417 plays an important role in stabilizing RBD-ACE2 and RBD-CB6 complexes through the formation of distinct salt bridges.
In an effort to further characterize these salt bridges, Luan and Huynh noted that K417 in the RBD interacts with more atoms in CB6 compared to ACE2. The salt bridge formed in RBD-ACE2 complexes are weakened due to electrostatic interactions inside the water-exposed salt bridge. Alternatively, the salt bridge formed in RBD-CB6 complexes is buried inside the protein and is very stable.
Achilles' heel of SARS-CoV-2
Molecular dynamics simulations with free energy calculations revealed that the K417N mutation significantly reduces the binding affinity between the RBD and CB6. The K417N mutation also reduces the binding affinity between the RBD and ACE2, but to a much lower degree than that between the RBD and CB6. The loss of binding affinity due to a single mutation in the South African variant makes it less competitive, in terms of ability to enter host cells, compared to wildtype SARS-CoV-2.
The study also showed that the K417N mutation allows the variant to escape from many human antibodies other than CB6 by removing a salt bridge buried in the RBD–antibody interface. This provides evidence that the K417N mutation developed as a result of viral adaptation to the human immune system or natural selection pressure from human neutralizing monoclonal antibodies. The K417N mutation seems to sacrifice its binding affinity for ACE2 to survive the attack of the antibodies, the authors explained.
While the K417N mutation causes reduced binding affinity, the researchers suggested that the gains in RBD-ACE2 binding resulting from both the N501Y and E484K mutations might be enough to compensate for the loss caused by the K417N mutation. Cumulatively, all three mutations found in the South African variant yield a stronger ACE2 interaction than that of the wildtype virus.
The insights from the study could help explain why the virus has selected for the K417N mutation during viral evolution and could aid scientists in the design of more efficacious monoclonal antibodies for the treatment of COVID-19 patients infected with new SARS-CoV-2 variants or vaccines to protect against SARS-CoV-2 variants of concern.
Do you have a unique perspective on your research related to infectious diseases or virology? Contact the editor today to learn more.
Copyright © 2021 scienceboard.net